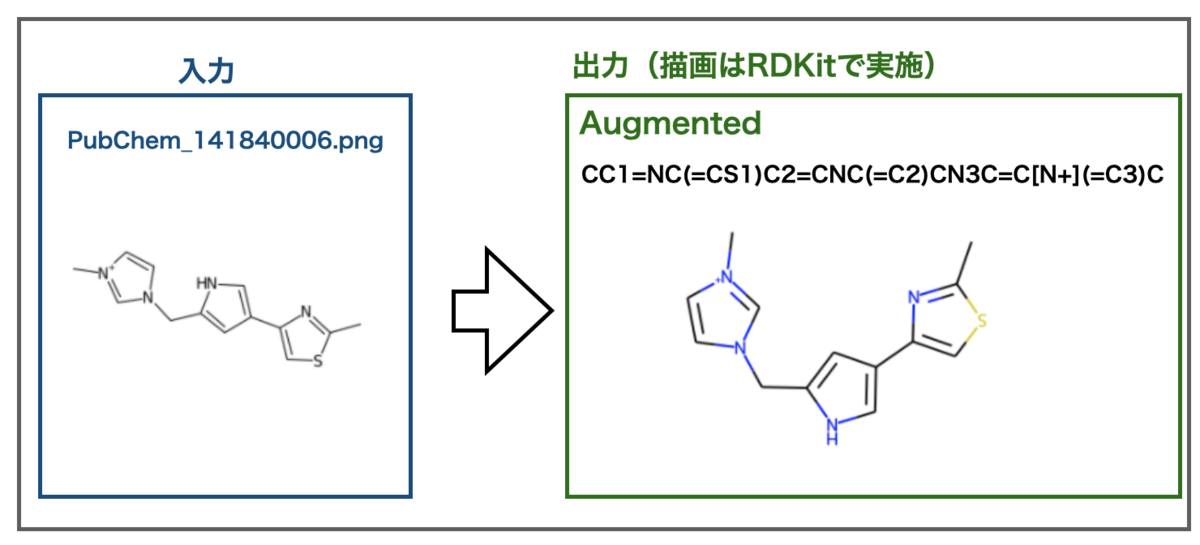
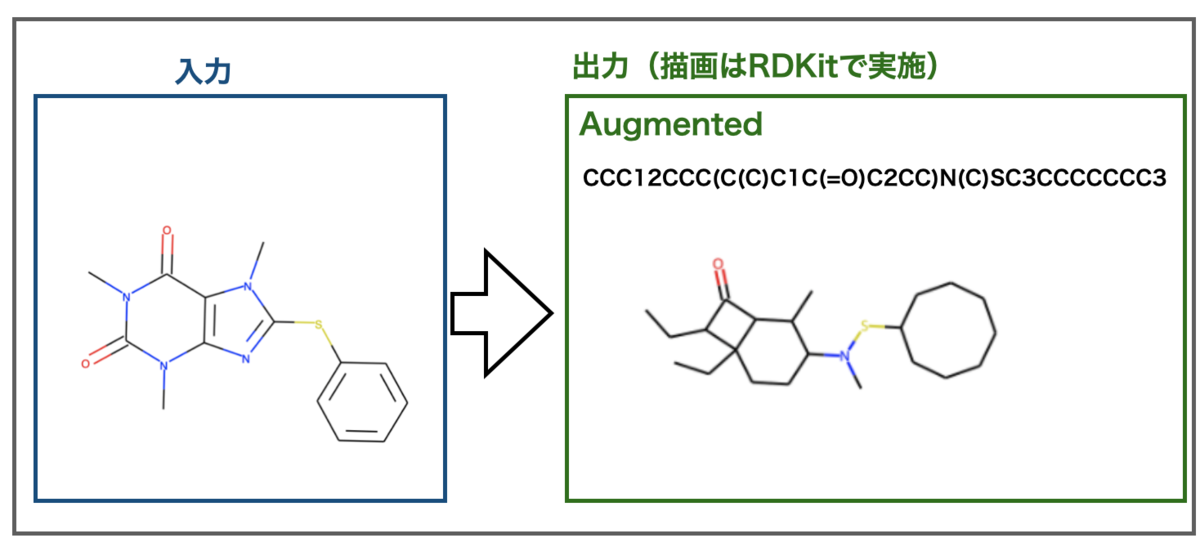
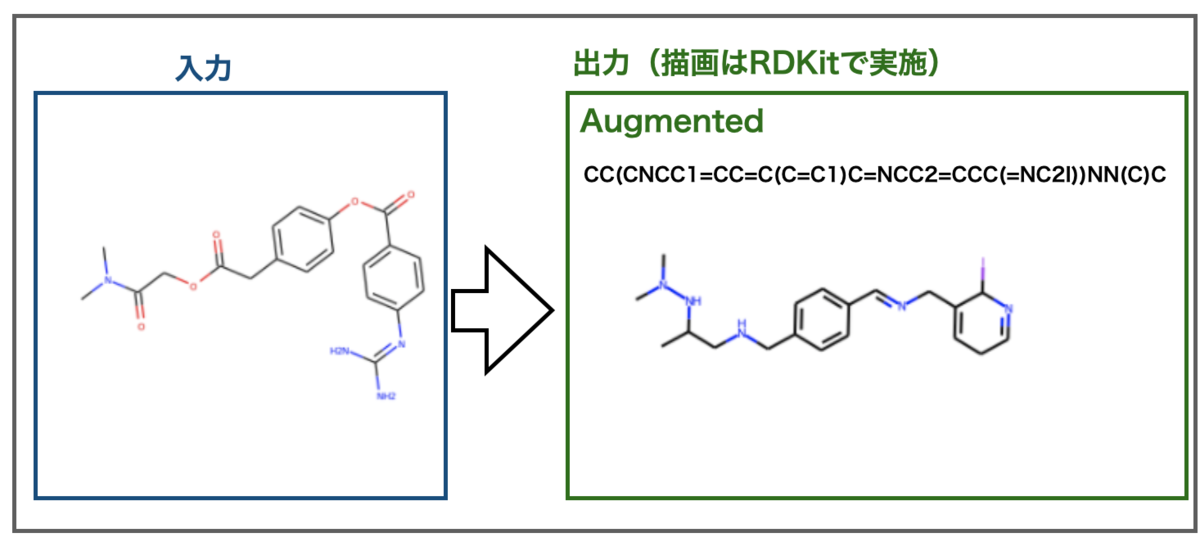
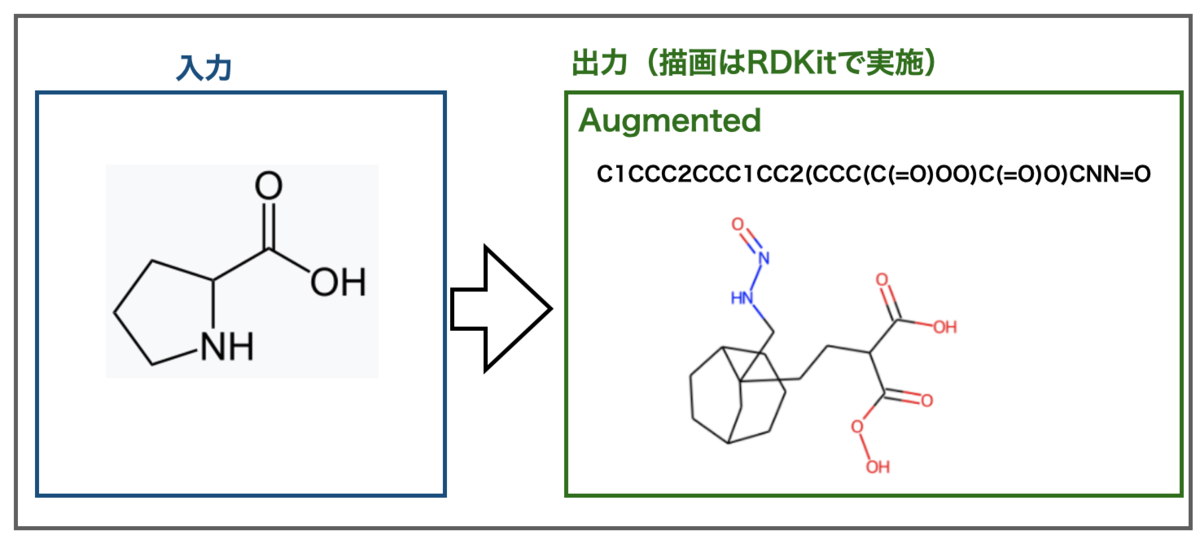
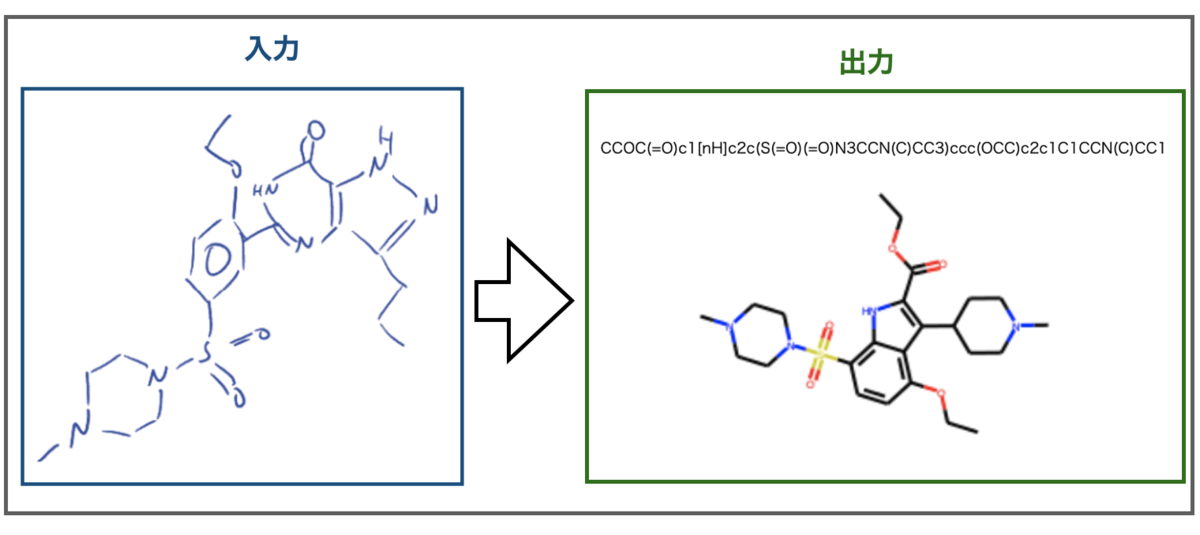
DeepMindのDFTのやつ(DM21)がColabで動きました
こんな感じでDM21がColab上で動かせました。コードはDM21のGitHubのREADMEから抜粋しました。
メタンの計算で14分かかりました。何が計算されたのかいまいち分かっていないです。
1. DM21のインストール
!pip install git+git://github.com/deepmind/deepmind-research.git#subdirectory=density_functional_approximation_dm21
2. 必要なライブラリのインポート
DM21はPySCFというソフトウェアに基づいて作られているので基本的にはPySCFと同じやり方で計算できるそうです。残念ながら私はPySCFを使った事がありません。
import density_functional_approximation_dm21 as dm21 from pyscf import gto from pyscf import dft
3. 計算したい分子の用意
メタン分子を構築するための情報はDM21のGitHubのREADMEを利用しています
# Create methane methane = gto.Mole() methane.atom = """H 0.000000000000 0.000000000000 0.000000000000 C 0.000000000000 0.000000000000 1.087900000000 H 1.025681956337 0.000000000000 1.450533333333 H -0.512840978169 0.888266630391 1.450533333333 H -0.512840978169 -0.888266630391 1.450533333333""" methane.basis = 'def2-qzvp' methane.verbose = 4 methane.build()
分子が構築できるとこんな結果がでます。
[CONFIG] conf_file None
[INPUT] verbose = 4
[INPUT] num. atoms = 5
[INPUT] num. electrons = 10
[INPUT] charge = 0
[INPUT] spin (= nelec alpha-beta = 2S) = 0
[INPUT] symmetry False subgroup None
[INPUT] Mole.unit = angstrom
[INPUT] 1 H 0.000000000000 0.000000000000 0.000000000000 AA 0.000000000000 0.000000000000 0.000000000000 Bohr
[INPUT] 2 C 0.000000000000 0.000000000000 1.087900000000 AA 0.000000000000 0.000000000000 2.055833050914 Bohr
[INPUT] 3 H 1.025681956337 0.000000000000 1.450533333333 AA 1.938257988385 0.000000000000 2.741110734552 Bohr
[INPUT] 4 H -0.512840978169 0.888266630391 1.450533333333 AA -0.969128994193 1.678580657029 2.741110734552 Bohr
[INPUT] 5 H -0.512840978169 -0.888266630391 1.450533333333 AA -0.969128994193 -1.678580657029 2.741110734552 Bohr
nuclear repulsion = 13.4613239153198
number of shells = 57
number of NR pGTOs = 209
number of NR cGTOs = 177
basis = def2-qzvp
ecp = {}
CPU time: 6.55
DFT計算するためのソルバーを作成
RKSはRestricted Kohn-Shamの略の様です。DM21汎関数をソルバーに代入することで、DM21を使った計算ができる様になります。
# Create a DFT solver and insert the DM21 functional into the solver.
mf = dft.RKS(methane)
mf._numint = dm21.NeuralNumInt(dm21.Functional.DM21)
DFT計算の実行
%%time
# Run the DFT calculation.
mf.kernel()
******** <class 'pyscf.dft.rks.RKS'> ********
method = RKS-RHF
initial guess = minao
damping factor = 0
level_shift factor = 0
DIIS = <class 'pyscf.scf.diis.CDIIS'>
diis_start_cycle = 1
diis_space = 8
SCF conv_tol = 1e-09
SCF conv_tol_grad = None
SCF max_cycles = 50
direct_scf = True
direct_scf_tol = 1e-13
chkfile to save SCF result = /content/tmpmc1ww10e
max_memory 4000 MB (current use 1094 MB)
XC library pyscf.dft.libxc version 5.1.7
S. Lehtola, C. Steigemann, M. J. Oliveira, and M. A. Marques, SoftwareX 7, 1 (2018)
XC functionals = LDA,VWN
P. A. M. Dirac, Math. Proc. Cambridge Philos. Soc. 26, 376 (1930)
F. Bloch, Z. Phys. 57, 545 (1929)
S. H. Vosko, L. Wilk, and M. Nusair, Can. J. Phys. 58, 1200 (1980)
radial grids:
Treutler-Ahlrichs [JCP 102, 346 (1995); DOI:10.1063/1.469408] (M4) radial grids
becke partition: Becke, JCP 88, 2547 (1988); DOI:10.1063/1.454033
pruning grids: <function nwchem_prune at 0x7fce59db0710>
grids dens level: 3
symmetrized grids: False
atomic radii adjust function: <function treutler_atomic_radii_adjust at 0x7fce59db0b90>
small_rho_cutoff = 1e-07
Set gradient conv threshold to 3.16228e-05
tot grids = 53998
関係なさそうな部分を省略します。SCFサイクルが走ってる感じの記述がずっと並んでました。
最終的な結果は以下
Extra cycle E= -40.5178537368252 delta_E= 4.76e-10 |g|= 5.27e-08 |ddm|= 6.94e-08
converged SCF energy = -40.5178537368252
CPU times: user 25min 26s, sys: 12.7 s, total: 25min 38s
Wall time: 13min 41s
収束したSCFエネルギーは「 -40.5178537368252」(単位はHartree?)で、計算に14分弱かかりました。
GitHubの計算結果は「'CH4': -40.51785372584538」だったのでほとんど同じ値が出ていそうです。
思ったより計算時間がかかりました。
DeepMindのDFT論文を読んだメモ
今年はDeepMindの論文祭りなのでしょうか?数学者とのコラボレーションがNatureに発表されたかと思いきや、今度はScienceにDFTに関する文献が出されたそうです。
数学ははなから諦めていますし、理論化学もさっぱりですが、やっぱりDFTの方はちょっと知りたい。
というわけでDFT論文、大体こんな話ですかー?っていう感想文です。

- 1. Science文献が公開されてた!
- 2. 前置き
- 3. 論文の中身
- 4. どうしてDeep Learningがよかったんだろう?
- 5. DFT x Deep Learningって他にもみたことあるけど?
- 6. おわりに
1. Science文献が公開されてた!
Scienceの文献自体はオープンアクセスではありませんが第一著者のJames Kirkpatrick博士がResearchgateにアップロードしてくださっています。
下記、DeepMindのブログからもリンクされているので正当な手続きで公開されているはず。。。*1
またOpen AccessのPDFがこちらから閲覧できます。こっちも上の記事からリンクされてました。
2. 前置き
2-1. 前置き① ~ DFTの未解決問題? ~
そもそもDFTにおける未解決問題とは何だったのでしょうか?
多分こういう話です。
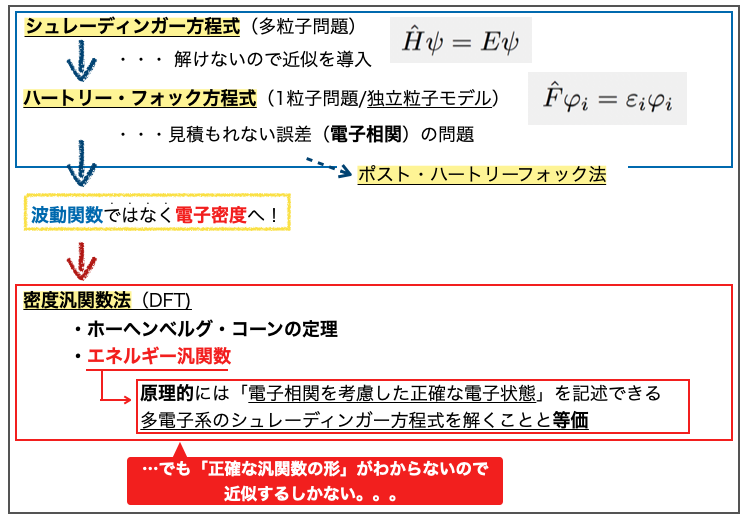
シュレーディンガー方程式はそのままでは解けないので近似の導入によりハートリー・フォック方程式を解く問題に変換されました。 それでも見積もれない誤差(電子相関 の問題)があるため様々なポスト・ハートリーフォック法が出てきました。
これに対して波動関数ではなく電子密度から出発しても系の状態(物理量)を知ることができるよ!そっち解こうぜ!ってのが密度汎関数法(DFT)のアプローチです。
ポスト・ハートリーフォック法ではさまざまな電子間の相互作用を考慮するため、電子数が多くなる(系が大きくなる)と計算量が膨大になり現実的に解くのが難しくなってしまうそうです。DFTは「より計算量が少なくて済むため現実的な時間で解くことができる」という利点がある様です。
ちょっとDFTに関する専門用語を貼ってそれっぽくします。

電子密度から系の状態がわかるというのはホーヘンベルグ・コーン定理に基づくそうです。この定理から電子密度とエネルギーの関係性を表すエネルギー汎関数というものがでてきます。
DFTではエネルギー汎関数に基づき原理的には「電子相関を考慮した正確な電子状態」を記述することができ、多電子系のシュレーディンガー方程式を解くのと等価になるそうです。
ここで問題となるのが「エネルギー汎関数の正確な形がわからない」ということで、これがDFTの未解決問題にあたるようです。
2-2. 前置き② ~ コーン・シャム近似と交換相関汎関数 ~
さて、エネルギー汎関数は正確な形がわからないため様々な近似モデルが提案されました。 初期のモデルでは精度が悪くて非実用的だったようですが、より改善したコーン・シャム近似が導入されたことでDFTが実用的なものになったそうです。
ざっとこんな感じみたいです。

コーン・シャム近似では、それ以前の近似モデルとは異なる新しい項、交換相関汎関数(exchange-correlation functional)が追加されています。
以前のモデルの精度の悪さは運動エネルギー汎関数の近似の悪さと考え、運動エネルギーを正確・計算可能な形に表したうえで、表現仕切れない残りを交換相関汎関数に押し込めています。また、交換相関汎関数には、電子間相互作用エネルギーのうち電子間クーロンエネルギーで近似できない残りも押し込められています。
よくわかりませんが、うまく表せない部分を押し込めて煮詰めた闇鍋部分が交換相関汎関数ってことですね(???)
3. 論文の中身
3-1. DeepMindのDFTモデル(DM21)
また前置きが長くなってしまいました。本題のDeepMindの文献についてです。
この論文ではDM21と名付けたDeepLearningに基づく汎関数(functional)が提案されています。
DFTでは「電子密度とエネルギーとの間に関係がある」ことがわかっていますが、その「エネルギー汎関数の正確な形がわからない」というのが大きな問題でした。
「未解決のブラックボックスな関数がある。。。それならデータから学習した最高精度のモデルで置き換えちゃえばいいじゃん!」って話ですね!・・・たぶん。
より具体的には標準的なコーン・シャム近似(Kohn-Sham DFT)のうち、交換相関項(exchange-correlation term)を学習させて置き換えています。先に見た闇鍋部分ですね!
学習の概要は論文のFig. 1をご参照いただくとして、大体こんな感じです。
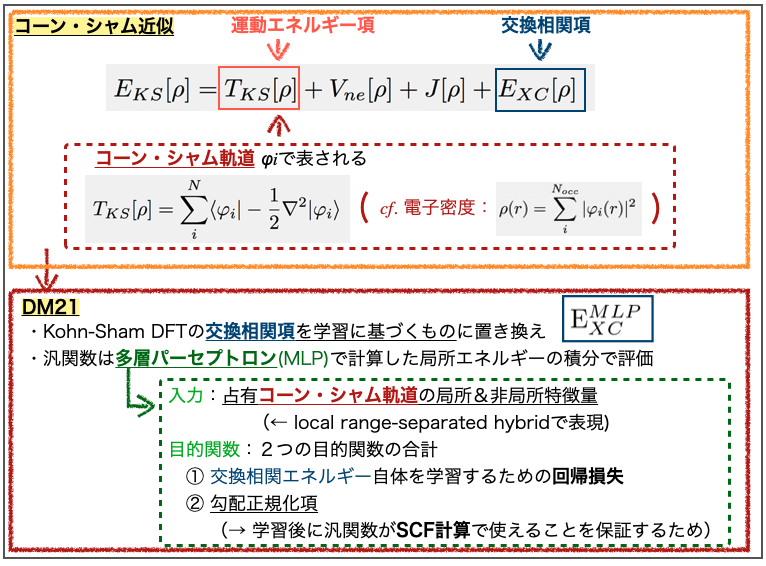
多層パーセプトロン(mulitlayer perceptron, MLP)の入力は占有されたコーン・シャム軌道(occupied Kohn-Sham orbitals)の局所的(local)特徴量と非局所的(nonlocal)特徴量です。コーン・シャム軌道はコーン・シャム近似で運動エネルギー項を表すために導入された波動関数に類似した形のものです。
この入力は「local range-separated hybrid」で表現できるそうですが、これが何かよくわかりませんでした。局所的な距離(r1, r2,…, rG)で区切ったものの組み合わせ(hybrid)という意味でしょうか??? お分かりの方ご教示いただけると嬉しいです。
また、学習の目的関数としては2つの目的関数の合計を用いています。ひとつは交換相関エネルギー自体を学習するための回帰損失(regression loss)で、もう一つは勾配正規化項(gradient regularization term)です。
前者はわかりやすいですが、後者は学習後の推論フェーズで重要になるようです。勾配正規化項を目的関数としておくことで、汎関数の導関数(derivative)が自己無撞着場の手続き(self-consistent filed (SCF) calculation)で利用できることが保証されるそうです。
DFTの計算ではコーン・シャム方程式を解くことが行われますが、この方程式は行列形式に置き換えられ固有値を求める問題となります。この固有値問題をとくための繰り返しの手続きがSCF計算と呼ばれるもののようです。
3-2. DM21が改善した2つの課題
さて、DeepMindの提唱するDFTモデル、DM21で行われていることがぼんやり見えてきました。ではDM21でどの様な問題が解消されたのか?より具体的にみてみます。
何より名は体を表す!
論文のタイトル「Pushing the frontiers of density functionals by solving the fractional electron problem」が表す通り「fractional electronの問題」です!・・・何のこっちゃ???
DeepMindのブログ記事によると、既存のDFT計算には長年解決できなかった2つの課題、①非局所化の誤り(the delocalization error)と②スピン対称性の破れ(spin symmetry breaking)があったそうです。
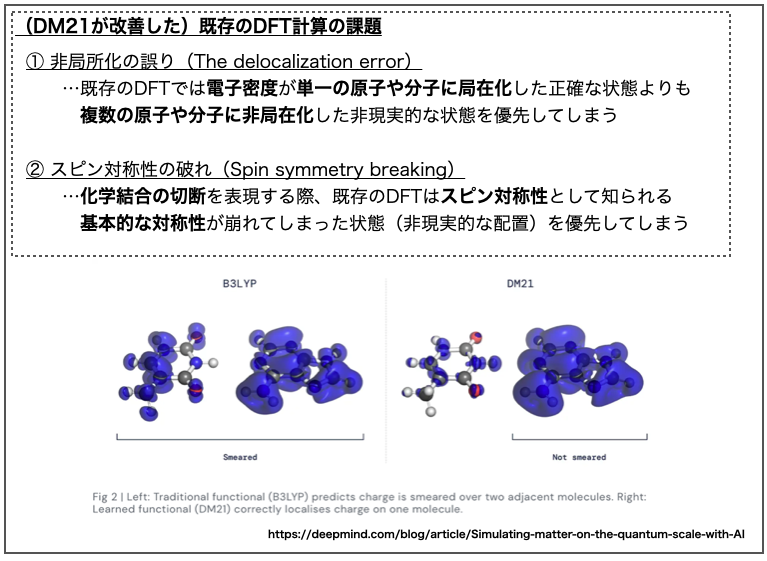
要するにDFTの既存の汎関数の近似モデルは不完全なので、「物理化学的に考えたら現実的にはこうだよね!」ってのと違う計算結果になってしまう、ってことみたいです。電子が1個、2個ではなく、分数個(fractional)あるとか言われても気持ち悪いですよねー。
ってことで、それぞれもう少し見てみます。
3-2-1. 非局在化の誤り
「① 非局所化の誤り」についてはDeepMindのブログ記事の図(Fig. 2)が分かりやすいので上に引用させていただきました。*2
上図ではDNAの塩基ペア(アデニン(A)-チミン(T))間における電荷の分布が計算されています。左が従来の手法(B3LYP)、右がDeepMindの手法(DM21)で計算した結果で、ブルーの網がけで電荷密度(charge density)が表現されています。
DNAには遺伝情報を伝えるという役割以外にも、電荷を輸送する(charge transport)という物理化学的にとても面白い性質が報告されています。2011年にはNature Chemistryにこんな論文も出ています。
この性質を理論的に理解しようとするとDFTの出番!となります。DNAの構成要素である塩基ペアの部位で「イオン化した状態で電荷がどの様に分布しているか?」知りたい、というモチベーションです。
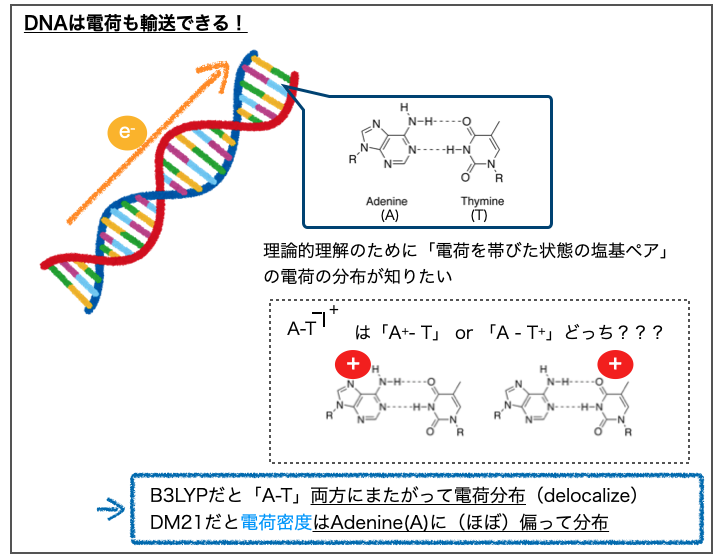
A-Tペアについてこの計算を行った結果が先に引用したFig. 2です。
アデニンがプラス電荷を帯びた「A+-T」の方が「A-T+」よりも安定というのが正解の様ですが、B3LYPでは(アデニンへの偏りはあるものの)A,T両方にまたがって電荷が分布しています。一方でDM21では(ほぼ)アデニンのみに電荷密度が偏った結果が得られています。
DFTにおいて「非局在化した計算結果が得られやすい」という課題が改善されていることの良いデモンストレーションになってますね。
3-2-3. スピン対称性の破れ
もう一方のDFTの課題「②スピン対称性の破れ」はそのままの話で「基本的には対称に配置されている方が安定のはずのスピンについて、対称性が崩れた結果をより安定と評価してしまう」ということの様です。
私では論文であげられている例を理解できなかったので、皆様Fig. 3B,3Cをご参照のうえ、ご教示いただけると嬉しいです。
なんかこんな感じの話。
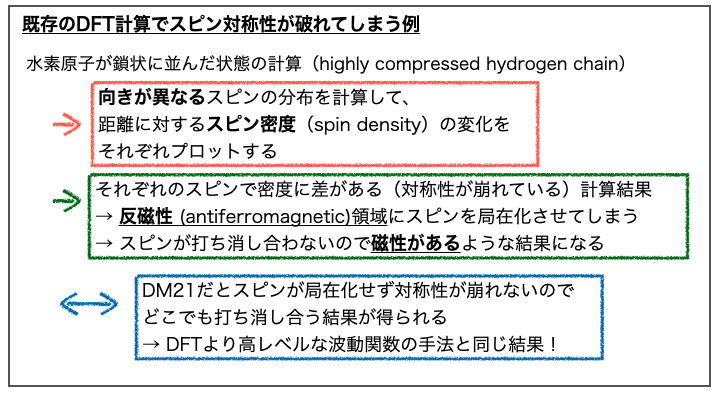
もうひとつあげられている例はビシクロブタンの異性化メカニズムの遷移状態についてでした。(こっちもよくわからなかった)
3-3. DM21のモデルの中身をもう少し ~ fractional chargeとfractional spin ~
DM21が解決しようとしたDFTの課題についても雰囲気がつかめてきました。では「何故、DM21のモデルでうまくいったのか?」。成功理由と関係のありそうなモデルのデザインの中身をもう少しみてみます。
DM21の取り組んだ課題は大きくは「fractional electronの問題」でした。この問題に対するため、DM21はfractional electronを有する系についての2つのタイプの制約条件、①fractional charge(FC)と②fractional spin(FS)に従う様にデザインされたそうです。
FCとFSはそれぞれ「非整数の総電荷(noninteger total charge)の系」と「非整数スピン磁性化(noninteger spin magnetization)の系」です。これらの系は当然、非現実的な架空のもの(fictitious)です。
ですが、「現実の電荷密度もFCやFSの特徴を持つ領域を含みうる」ので、この「理想状態の問題を正確にモデル化」することができれば、「多様な分子や素材についても上手く機能する汎関数」が得られるのではないか?という考えに基づくようです。
現実には無い架空の特性を持つユニットをモデルにくみこむあたりは、AlphaFold2でアミノ酸残基をガスとしてモデル化してフィッティングしていた解き方に通じるところがあって面白いです。
さて、fractional electronの問題を解消するために使われたFCとFSですが、これらの制約条件はデータで表現する事ができるため、これに従って分子系のエネルギーを再現する様に汎関数を学習すれば良い、ということになるらしく、Deep Learningと相性の良い問題に帰着できたことになったようです。
手法とモデル化がうまく合致していてすごい!
3-4. fractional chargeとfractional spinをもう少し
架空のモデル、fractional charge (FC) とfractional spin (FS) ですが、これらはDM21で初めて導入されたものではなく、既存のDFT汎関数でも使われていました。ただし、従来の汎関数ではFCとFSに関連したエラーが解消できず問題となっていた様です。
このエラーは①電荷を帯びた分子と、②閉殻中性分子(closed-shell neutral molecules)において結合の切断の表現が不正確になってしまう、という問題です。
詳細は論文のFig. 2をご参照いただくとして、大体こんな話です。

それぞれ、①の問題はDFTの「電子密度の非局在化」の課題、②はDFTの「スピン対称性の破れ」の課題と関連していることが何となく分かりますね。
これらのFCとFSに関する問題について、DM21は従来の汎関数を上回るパフォーマンスが得られたので、先にみた実課題での検証(A-T, hydrogen chain, etc.)に進んだ、というのが論文の流れです。*3
以上が、DeepMindのデザインしたDFT汎関数DM21の概要と、その解決しようとした課題についてでした。
論文についてはここまでで、あとは雑感です。
4. どうしてDeep Learningがよかったんだろう?
さて、今回もDeepMindはDeep Learningで分野の壁を突き破ったわけですが、何故DFTの課題にDeep Learningを適用するのがよかったのでしょうか?
今回の論文を受けたNatureのNews記事(DeepMind AI tackles one of chemistry’s most valuable techniques)から引用します。
“It’s sort of the ideal problem for machine learning: you know the answer, but not the formula you want to apply,” says Aron Cohen, a theoretical chemist who has long worked on DFT and who is now at DeepMind. (Nature 600, 371 (2021))
「答えがわかっていて、わかっていない式の形を求めるのは機械学習にとって理想的な問題」だそうです。
この考えの背景にあるのはおそらく「普遍近似定理(universal approximation theorem)」ではないでしょうか?

上記は2層についての定理ですが、さらに多層にすることの利点は層の深さに応じて表現力が増す、適応力があがるといったことにある様です。
詳細は上記の書籍や、東京大学大学院 鈴木大慈先生がSlideShareに公開してくださっている資料(深層学習の数理)などをご参照ください。
雑な理解ですが、多層のニューラルネットワークであれば「任意の関数を精度よく近似」できるので、「関数の形がわかっていない問題」にDeep Learningを適応していい感じにすれば「良い近似モデルが得られる!」ってことになりそうです。
5. DFT x Deep Learningって他にもみたことあるけど?
最後に何故この研究にインパクトがあったのか?ちょっと想像してみます。
というのも「DFTに対してDeep Learningを適用して改善した!」という論文・ニュース記事などは偶に目にするので何が違うのかなー?と気になったからです。
例えばこういうのです。
どちらもニューラル・ネットワーク・ポテンシャル(NNP)の研究です。
一つ目はフロリダ大学A.E. Roitberg教授らのグループによるもの(Chem. Sci., 2017,8, 3192-3203)で、2つめはPreferred Computational Chemistry社の提供するプラットフォームMatlantisに関する論文です(arxiv:2106.14583)。
私自身が専門分野外のため、これらの論文を比較対象として引用するのは間違っているかもしれません。ごめんなさい。
DFT計算はシュレーディンガー方程式を扱うよりも計算コストが低いという利点がありますが、それでも系が大きくなるにつれて必要な計算量が増え、非常に時間とコストのかかるようになってしまうようです。「計算のシミュレーションに時間かかるなら、実際に実験した方が速くない??」って言われそうです。
この課題があるため、Deep Learningで同等の計算結果が出せる様に学習し、計算時間のかかる部分を置き換えることで加速・コストを下げよう!というのがNNPの取り組みの様です。
何となくですが、これらの研究は計算コストの改善に焦点があり、既存のDFTが抱える精度上の課題の解決とはなっていなさそうです(たぶん)。
DM21は汎関数自体を作って、根本的な課題に取り組んだ、と言う点にインパクトがあったのかな?と思います。
6. おわりに
以上、今回はDeepMindのDFTに関するScience文献を読んでみた感想文でした。
量子化学に疎く、DFTの計算もやった事がなかったのでそもそも既存の汎関数に課題があることも知りませんでした。というより、論文を読み終えた今でもいまいち課題を理解しきれていなかったりします。
DFTの課題でスピン対称性の破れが上がっていたのに、Fractional spinのところではスピン非局在化の過剰評価が問題になっていて、結局対称性が破れるのと破れないのどっちが問題なの???となってしまいました。私に量子化学は難しすぎます。
なんとなくですが、適切なデータがある問題であれば、Deep Learningでうまく関係性を記述するモデルを作る事ができそうだ、ということが分かったのはよかったです。DeepMindが次にどんな課題に取り組むのか?楽しみですね!
ところで今回の論文のDM21ですが、素晴らしいことにGitHubで公開されています。ぜひぜひ専門家の皆様の使ってみた感想がお聞きしたいところです。
なお、今回の記事の前置き部分などは以前に書いたブログ記事を使いまわしています。
ずっと雰囲気から成長できていなくてすみません。今回も色々と間違いが多そうなのでご指摘いただけると嬉しいです。
ではでは!
アラート ≠ 禁止 〜ZINCデータベースの医薬品にPAINSフィルターをかけてみた話〜
創薬(dry)アドベントカレンダーを埋めてみます。アドベントカレンダーの記事を見ると「いよいよ年末かー」となりますね。
さて、今年も色々な格好いい分子が出てきてケミカルスペースは広大だなぁと思いました。
というわけで発表します!びっくりモルキュールオブザイヤー!
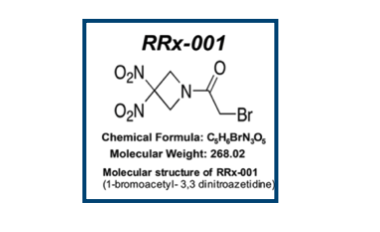
抗がん剤として臨床開発中だそうです。論文は以下。
- Discovery of RRx-001, a Myc and CD47 Downregulating Small Molecule with Tumor Targeted Cytotoxicity and Healthy Tissue Cytoprotective Properties in Clinical Development
J. Med. Chem. 2021(64)7261
ニトロ基2つにアミドのα位にハロゲン原子。。。見ただけで涙がちょちょ切れそうです。
これを臨床開発するなんて本当にびっくりですが、逆にいうと常識(?)みたいなものに囚われて有用性が見過ごされている分子がまだまだあるかもしれません。
そこで今回は「承認済みの医薬品の中にどれくらい忌避構造(アラート構造)が含まれているか?」調べてみることにしました。
1. この記事でやること
やることは単純です。ZINC15データベースにあるFDA approved drugsの一覧を使って、RDKitでPAINSフィルターにかかる分子がどれくらいあるか探してみるだけです。
2. データの準備
「ZINCデータベースは、バーチャルスクリーニングのために特別に準備された、市販の化学物質に関する精選されたコレクション」だそうです(Wikipedia - ZINCデータベースより)。
カリフォルニア大学サンフランシスコ校(UCSF)のJohn J. Irwin博士の研究室とBrian K. Schoichet博士の研究室により開発・維持されているそうで、現在はZINC20です。
- ZINC20—A Free Ultralarge-Scale Chemical Database for Ligand Discovery
J. Chem. Inf. Model 2020(60)6065
サーバーが混んでいてアクセスできなかったので今回はZINC 15を使います。
- ZINC 15 – Ligand Discovery for Everyone
J. Chem. Inf. Model 2015(55)2324)
よく知らないけどよく見るからこれを使っとけば良いんでしょう!知らんけど。
こんなページです。
ここにSubstancesがあるじゃろ。これをこうして。こうじゃ。
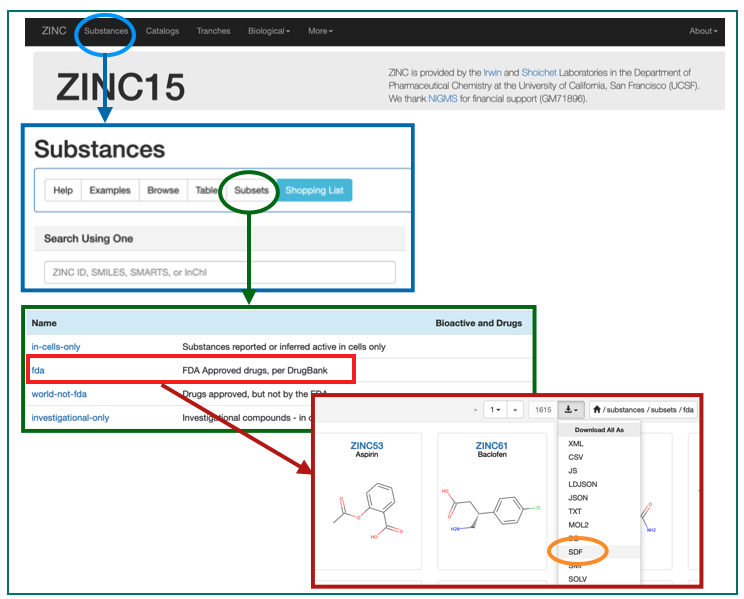
これでFDA承認済みの医薬品構造のSDFが手に入りました!
2015年に公開されたデータベースのようなので、少し古いかもしれませんが今回はこれで。。。
3. PAINS
3-1. PAINSフィルターって?
PAINSフィルターについては「化学の新しいカタチ」さんのブログ(RDKitのPAINSフィルターで化合物をスクリーニング)でわかりやすく解説してくださっています(RDKitのコードも!) 。
PAINSはPan-Assay INterference compoundsの略だそうです。
- New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays
J. Med. Chem. 2010(53)2719
ある種の部分構造を持つ化合物は、ハイスループットスクリーニング(HTS)を実施した際に、色々なアッセイ(pan-assay)で活性化合物としてヒットしてくるそうです。このような化合物は偽陽性である可能性が高くなるため要注意です。
そんなアッセイの妨害になる化合物(interference compounds)をフィルタリングして取り除けるように、「予め要注意な部分構造をリスト化しておこう!」というわけで作られたのがPAINSフィルターみたいです。
3-2. PAINSフィルターをもう少し
PAINSフィルターの具体的な中身についてもう少し見てみましょう!
2018年 ACS Chemical BiologyにPAINS公開後7年を経て再度その内容を振り返るといった趣旨の文献が出されています。こちらはオープンアクセスとなっています*2
- Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations
ACS Chem. Biol. 2018(13)36
「PAINSは数秒で簡単に100個も1000個も大量に化合物選別できて便利だけどブラックボックスのまま使うのはやめてね!」ってな感じです。
そもそもPAINSに上げられている構造はどうしてアッセイの邪魔になってしまうでしょうか? 文献では以下の様な特徴が指摘されています。

タンパク質や試薬と反応してしまう構造(求電子剤、光反応剤)は非特異的な不可逆反応を起こしそうなのでわかりやすいですが、キレート剤やフォトクロミズムといったものは考えてもみませんでした。確かにアッセイの原理と相性が悪そうです。なるほど!
さてPAINSフィルターをブラックボックスとしないために把握しておくべき注意点ですが、PAINSが「商用化合物を使ってPPI(protein-protein interaction)阻害を対象に行われた6つのHTS(AlphaScreen)における観察」に基づくものであることに由来する様です。*3
注意点はこんな感じ。

PAINSフィルターを使うときは用法・用量を守って適切に!
参考までに②のそもそもライブラリに含まれていなかった構造の例としてFig. 1を引用しておきます。

ついでにFig. 2も面白いので引用しておきます。メカニズムも用語も色々あってわかりにくいから整理して分類したよ!って図です。
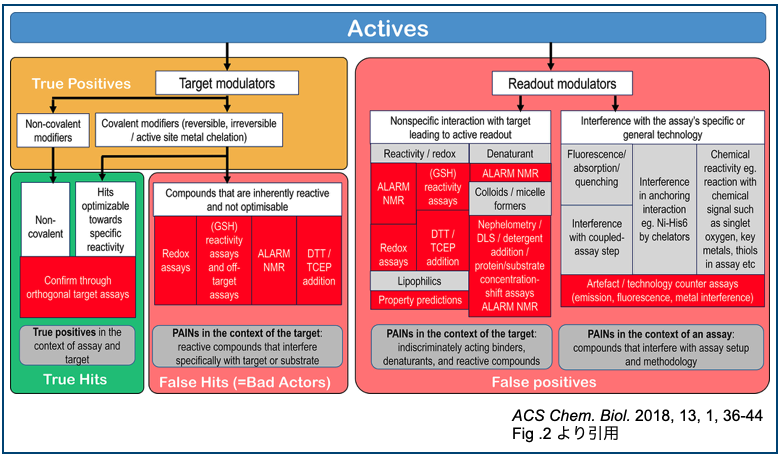
promiscuous binderとかfrequent hitter、bad actorってさらっと言えたら業界人っぽくないですか?・・・そうでもないですか。すみません。
4. RDKitのPAINSフィルターをつかってみよう
4-1. 簡単な使い方 ~ RRx-001を例に ~
だいたいPAINSのやりたいことがわかったので早速使ってみましょう!
まずは記事冒頭にあげたRRx-001を例にRDKitのPAINSフィルターを使ってみます。コードは化学の新しいカタチさんの記事とRDKitのブログ記事を参考にさせていただいています。
とりあえず化合物構造を準備します。
from rdkit import Chem from rdkit.Chem import AllChem, Draw # PubChemよりRRx-001のSMILESを取得(PubChem ID : 15950826) RRx_001_smiles = "C1C(CN1C(=O)CBr)([N+](=O)[O-])[N+](=O)[O-]" # Molオブジェクトを作って描画 RRx_001_mol = Chem.MolFromSmiles(RRx_001_smiles) Draw.MolToImage(RRx_001_mol)
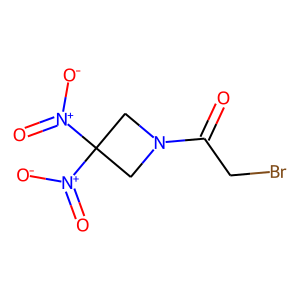
RDKitのPAINSフィルターには4つ(PAINS、PAINS_A、PAINS_B、PAINS_C)あります。ABCはそれぞれPAINSを作る際のHTSで150以上の化合物に含まれた構造(Family_Filter_A)、15 - 149化合物(Family_Filter_B)、14以下(Family_Filter_B)に対応しているようです。
よくわからないので「PAINS(無印)」を使ってみす。
FilterCatalogに実装されており、FilterCatalogParamsオブジェクトに使いたいフィルタリング要素を追加してフィルターを作った後、目的の化合物に適用すれば良いそうです。
# インポート from rdkit.Chem import FilterCatalog # パラメータのオブジェクトを作る param = FilterCatalog.FilterCatalogParams() # PAINSフィルターをパラメータに追加する param.AddCatalog(FilterCatalog.FilterCatalogParams.FilterCatalogs.PAINS) # フィルターを作成 filt = FilterCatalog.FilterCatalog(param)
フィルターができたので適用します。
HasMatchで「フィルター中の部分構造が含まれているかどうか?」を確認することができます。
print(filt.HasMatch(RRx_001_mol)) # False
False !!! RRx-001はPAINSフィルターにはかからない化合物でした!見た感じヤバいのに。。。
他のフィルターではどうでしょうか?RDKitには他に3つのフィルター(BRENK、NIH、ZINC)が用意されています。それぞれ適用してみましょう。
# フィルター名のリスト filter_names = ["BRENK", "NIH", "ZINC"] # 順番に適用 for name in filter_names: p = FilterCatalog.FilterCatalogParams() fc = "FilterCatalog.FilterCatalogParams.FilterCatalogs." + name p.AddCatalog(eval(fc)) f = FilterCatalog.FilterCatalog(p) res = f.HasMatch(RRx_001_mol) print("Does filter " + name + " detect RRx-001? : " + str(res) ) # Does filter BRENK detect RRx-001? : True # Does filter NIH detect RRx-001? : True # Does filter ZINC detect RRx-001? : False
BRENKとNIHではRRx-001はフィルターにかかりました!どの構造が認識されたのでしょうか?
GetMatchesで「具体的にどのようなフィルターにひっかかったか?」取り出せるそうです。NIHを例に行ってみます。
# NIHフィルターを作成 p2 = FilterCatalog.FilterCatalogParams() p2.AddCatalog(FilterCatalog.FilterCatalogParams.FilterCatalogs.NIH) f2 = FilterCatalog.FilterCatalog(p2) # フィルターに認識された構造情報を取り出す matches = f2.GetMatches(RRx_001_mol) for match in matches: print(match.GetProp("description")) print(match.GetProp("Reference")) print(match.GetProp("Scope")) print("------------") """ alpha_halo_carbonyl Jadhav A, et al. Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease. J Med Chem 53 (2009) 37D51. doi:10.1021/jm901070c. annotate compounds with problematic functional groups ------------ non_ring_ketal Jadhav A, et al. Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease. J Med Chem 53 (2009) 37D51. doi:10.1021/jm901070c. annotate compounds with problematic functional groups ------------ primary_halide_sulfate Jadhav A, et al. Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease. J Med Chem 53 (2009) 37D51. doi:10.1021/jm901070c. annotate compounds with problematic functional groups ------------ """
「alpha_halo_carbonyl、non_ring_ketal、primary_halide_sulfate」の3つの部分構造が認識されること、NIHフィルターが以下の文献をもとにしていることがわかりました。
- Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease
J. Med. Chem. 2010 (53)37
このままではわかりにくいので構造式上で確認してみましょう。フィルターにかかった構造のアトム番号はGetFilterMatchesで取り出せるそうです。
# descriptionのリスト descriptions = [match.GetProp("description") for match in matches] # アトム番号のリストのリスト atom_nums_list = [] for match in matches: atom_nums = [x[1] for x in match.GetFilterMatches(RRx_001_mol)[0].atomPairs] atom_nums_list.append(atom_nums) # 描画 Draw.MolsToGridImage([RRx_001_mol for _ in range(len(matches))], highlightAtomLists=atom_nums_list, legends=descriptions)
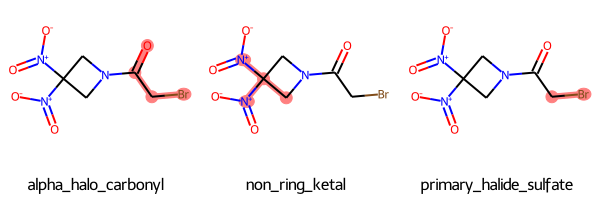
構造を描画するとわかりやすいですね!臭素原子はカルボニルα位のハロゲンや、primary-halideとして認識されています。意外にも(?)、ニトロ基はニトロ基としてではなく、同一炭素原子に2つの窒素原子が結合した構造(non_ring_ketal)として認識されている様です。
同じことの繰り返しなので省略しますがBRENKフィルターでは以下の結果が得られました。
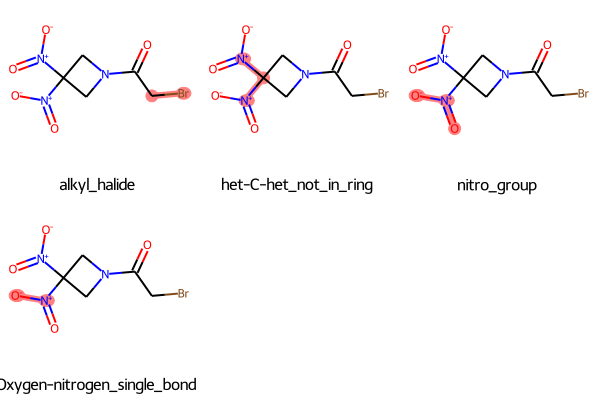
今度はニトロ基はnitro_group、Oxygen-nitrogen_single_bondとしても認識されている様です。見比べると面白いですね!
4-2. いざ承認化合物に適用!
RDKitのPAINSフィルターの使い方が大体わかりました。
それではようやく本題!ZINCデータベースから取り出したFDA承認化合物に適用してみましょう!
フィルターとしてはPAINSを使い、それぞれの化合物に「フィルターにマッチする部分構造がいくつ含まれているか?」検証してみます。
化合物数が多いのでPandasToolsでDataFrameとして扱います。
from rdkit.Chem import PandasTools import pandas as pd # SDFをデータフレームに読み込む df = PandasTools.LoadSDF("fda.sdf") print(df.shape) # (1615, 4) df.head(2)
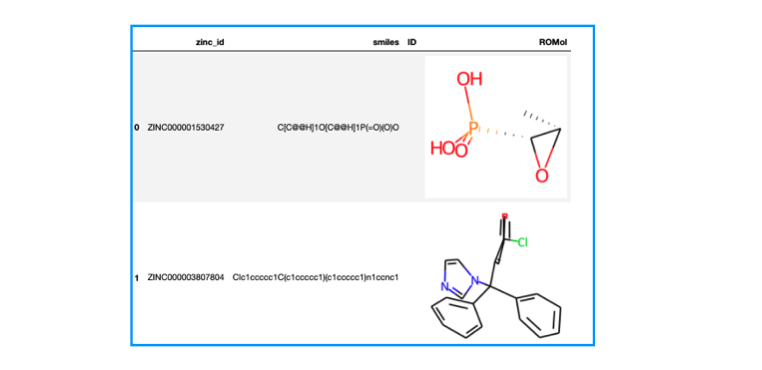
1615個の化合物がありました。zinc_id、smilesといったプロパティが含まれており、化合物は3次元に起こされた座標が入っている様です。
PAINSフィルターとマッチする構造を数える関数を作ってmap()でDataFrameに適用します。
# PAINSフィルター作り直し param = FilterCatalog.FilterCatalogParams() param.AddCatalog(FilterCatalog.FilterCatalogParams.FilterCatalogs.PAINS) filt = FilterCatalog.FilterCatalog(param) # フィルターにかかった数を数える関数 def match_num(mol): if filt.HasMatch(mol): matches = filt.GetMatches(mol) num = len(matches) else: num = 0 return num # PAINSフィルターにかかる化合物か否か?(True or False) df['PAINS'] = df.ROMol.map(filt.HasMatch) # フィルターにかかった回数は? df['filter_match_num'] = df.ROMol.map(match_num)
そもそもFDA承認化合物にPAINSフィルターにかかったものはあったのでしょうか?
# PAINSフィルターでTrueになった化合物の数 PAINS_drugs_num = df['PAINS'].sum() print("PAINS detected {} compounds.".format(PAINS_drugs_num)) # PAINS detected 79 compounds.
# 割合 PAINS_ratio = round((PAINS_drugs_num / len(df))*100) print("ratio : {} %".format(PAINS_ratio)) #ratio : 5 %
79化合物がPAINSフィルターにひっかかることがわかりました!
承認済み1615化合物の約5% にあたります。結構多いですね。
フィルターにかかった回数はどうでしょうか?
print(df['filter_match_num'].value_counts()) """ 0 1536 1 72 2 5 3 2 Name: filter_match_num, dtype: int64 """
72化合物で1回、5化合物で2回、2化合物で3回ひっかかっているようです。
3回かかった2化合物を取り出してみましょう。
df[df['filter_match_num'] == 3]
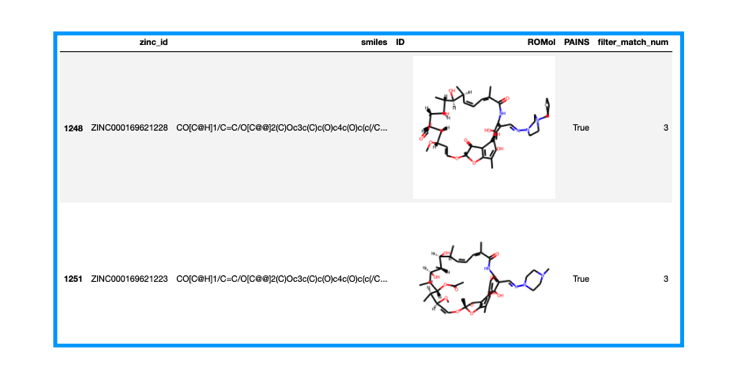
3次元だとよくわからないので2次元に描画しなおしてみます。マクロサイクルの化合物みたいなのでSetPreferCoordGenで描画の調整を行います。
# 該当化合物のSMILESを取り出してMolオブジェクトを作りなおす df3 = df[df['filter_match_num'] == 3] mols = [Chem.MolFromSmiles(smi) for smi in df3["smiles"]] # ZINC idを取り出す zinc_ids = [zinc_id for zinc_id in df3["zinc_id"] ] # マクロサイクルの描画を綺麗にする設定 Draw.rdDepictor.SetPreferCoordGen(True) # 描画 Draw.MolsToGridImage(mols, legends = zinc_ids)
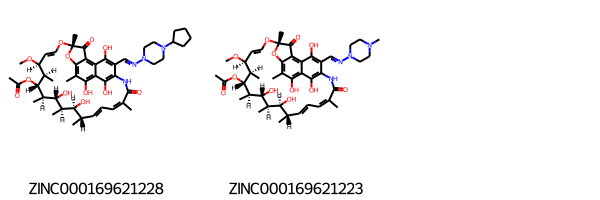
PubChemで調べたところ、それぞれCyclopentylrifampicinとRifampicineのようです。
大体同じ構造なのでRifampicineでPAINSフィルターに合致した構造をみてみます。コードは繰り返しなので省略します。
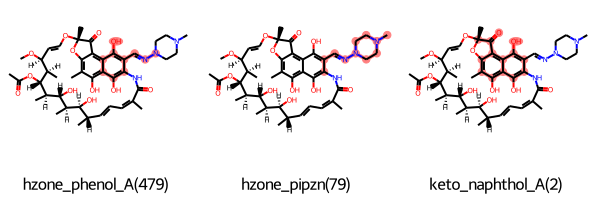
最初の二つはヒドラゾン構造とその周辺が認識されている様です。もう一つはケトンと共役したナフトール構造でした。
二重結合と共役したアミド構造やビニルエーテルなど他にも気になる構造はありますがこれらは認識されていない様ですね。
5. おわりに
以上、今回は見た目のインパクトが強いRRx-001を題材にRDKitのPAINSフィルターの使い方をお勉強し、ついでにZINC15から取得したFDA承認済み化合物にも適用してみました。
途中で引用したACS Chemical Biology「PAINS振り返り論文」中に、「PAINSは網羅的なものではない」という注意点が指摘されていました。実際に、RRx-001はPAINSには認識されないものの、他のフィルター(BRENK、NIH)ではアラートとして認識されていました。
また、FDA承認済み化合物とPAINSの関係についても上記文献に以下の記載がありました。
A small proportion (ca. 5%) of FDA-approved drugs contain PAINS-recognized substructures, these comprising both natural products and synthetic drugs.
ZINC15の例でも確かに5% でフィルターに引っかかりました。中でも3回フィルターにひっかかった化合物はRifampicineとCyclopentylrifampicin(リファペンチン)でした。
この2つは抗結核薬で、放線菌から取り出されたリファマイシンをリード化合物としたリファマイシン系化合物に分類されるもののようです。確かにいかにも天然物由来な構造です。
天然物由来の化合物には部分構造だけを取り出してみたら「そんな構造大丈夫なの?」と不安になるものが他にも多そうです。人間がデザインしたライブラリには絶対に入らなさそうな構造に有用性が見出されるのが天然物の面白さですよね!
先入観にとらわれなければライブラリにも自然界にもまだまだお宝が眠っているかもしれません。フィルターは適切に使いたいですね!
ところで、昨年に引き続き今年も対面で人に会うのが難しい一年でした。
来年は会えん 痛みがなくなるといいですね!
おしまい!
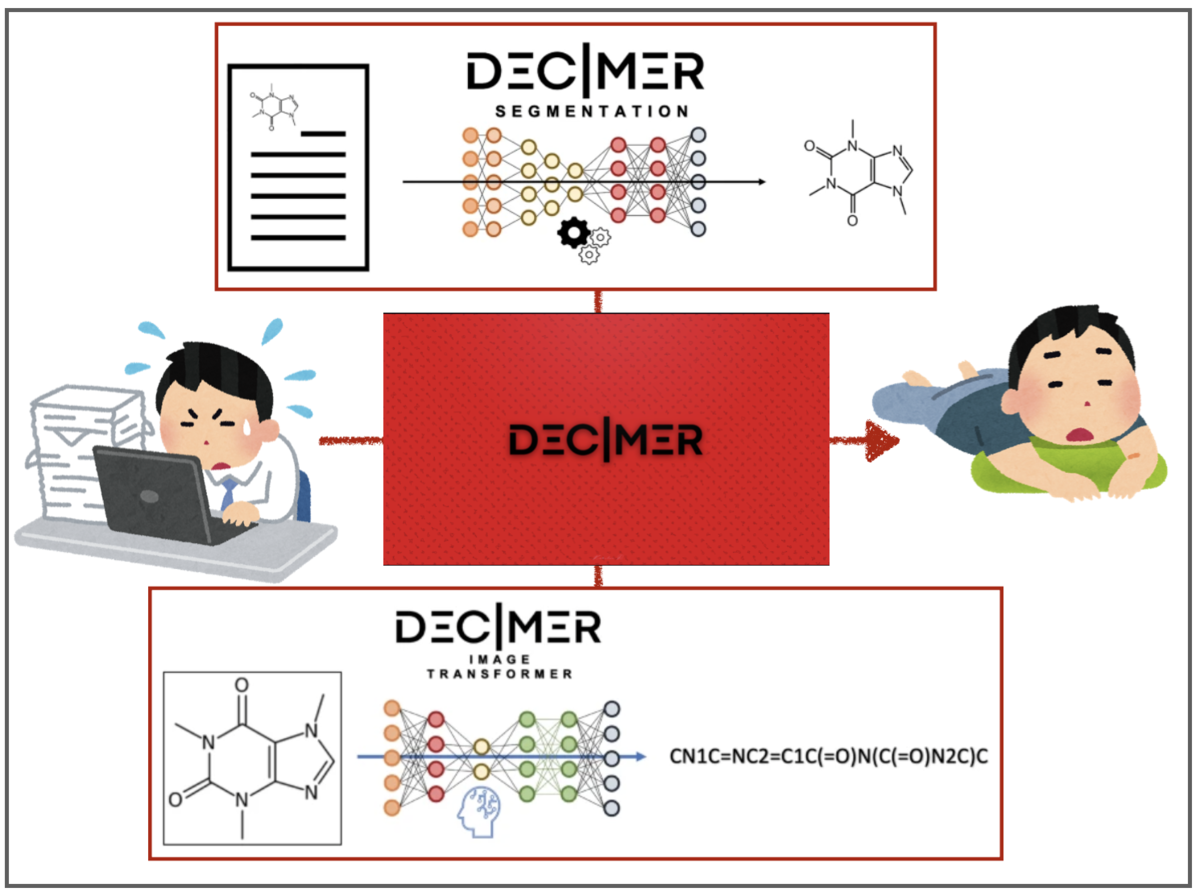
化学構造認識(OCSR) DECIMERプロジェクトとDECIMER 1.0で遊んだ話
前回の記事で化学構造式のOCRソフトImg2Molで遊んでみました。「画像に含まれる化学構造式を認識してSMILESに変換」してくれるというものでした。
Img2Molと同時期に同様のタスクを扱うソフト DECIMER 1.0が公開されています。
- 文献①(オープンアクセス) : DECIMER 1.0: deep learning for chemical image recognition using transformers J. Cheminform. 2021(13)61
- GitHub : DECIMAR-Image_Transformer
こちらもコードや学習済みパラメータを公開してくださっています‼︎ Img2Molとの比較も兼ねて遊んでみましょう!
1. DECIMER文献の流し読み
1-1. DECIMERプロジェクトについて
さて、Img2Molは製薬会社バイエルの機械学習チームによるものでしたが、DECIMERはドイツのFriedrich-Schiller大学Chirstoph Steinbeck教授の研究室(HPはこちら)で開発されたアカデミックな研究に由来するものです。
こちらもディープラーニングによる手法で、DECIMERはDeep lEarnig for Chemical ImagE Recognitionの略です。
このソフトは「化学構造式の光学認識(Optical Chemical Structure Recognition, OCSR)」の課題について、「最新の人工知能技術を使ったオープンソースの自動化されたソフトウェアを開発しよう!」というDECIMERプロジェクトの中の一つです(プロジェクトWebサイト)。
関連する文献として、今回取り上げる文献の前のバージョン(DECIMER(無印))や、、、
- 文献②:DECIMER: towards deep learning for chemical image recognition J. Cheminform. 2020(12)65
テキストと画像の入り混じった文献を区分けして、画像部分をとりだすセグメンテーションのためのツール、、、
- 文献③:DECIMER-Segmentation: Automated extraction of chemical structure depictions from scientific literature J. Cheminform. 2021(13)20
があります(どちらもオープンアクセス)。後者のツールはウェブアプリとして利用可能になっています。
全て筆頭著者はKohulan Rajanさんとなっていますので、こちらの方が中心になって進められているようですね。すごい。
1-2. DECIMER 1.0のモデル
それではDECIMER 1.0の具体的な中身を見てみましょう!Img2Molと同じくディープラーニングに基づく手法ですが、具体的な中身は結構違います。
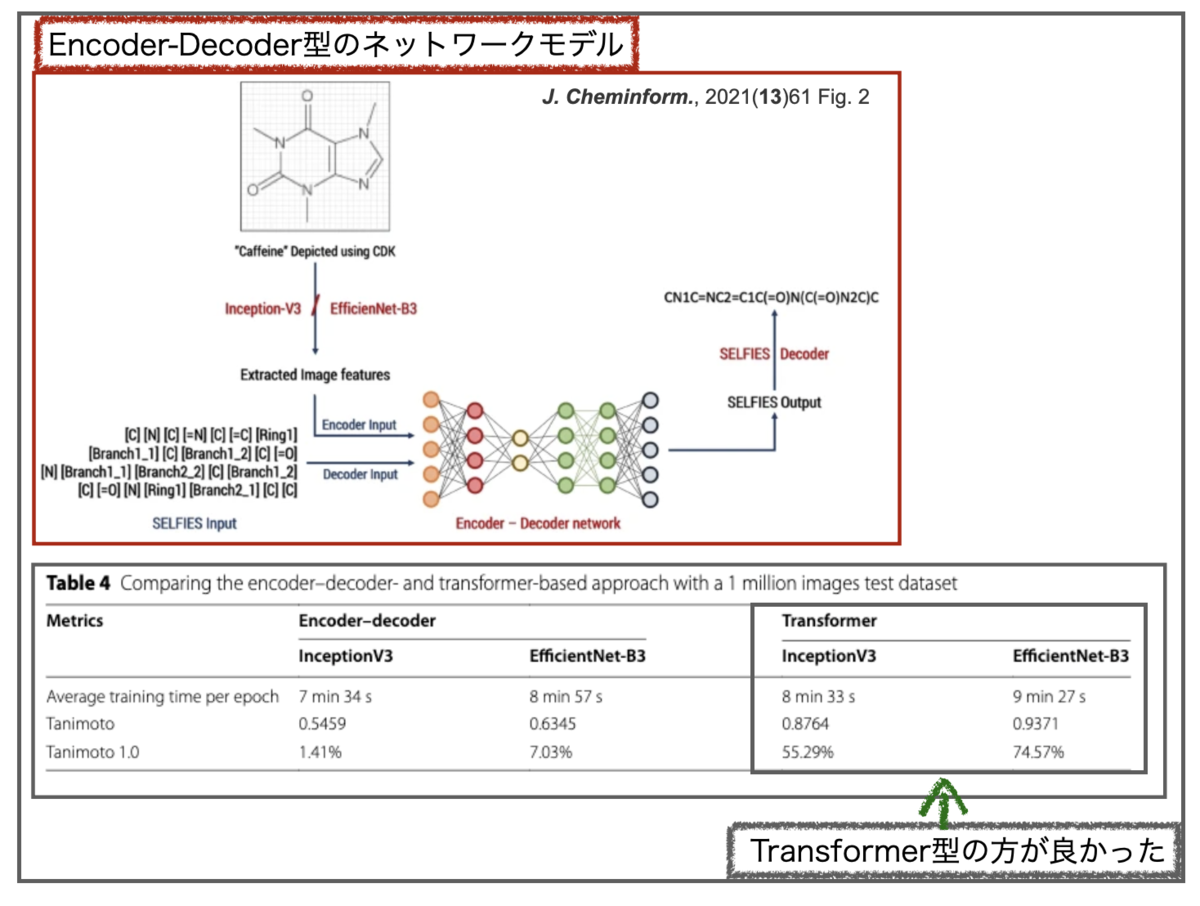
おさらいですが、Img2Molは画像を畳み込みニューラルネットワーク(CNN)で認識し、化学構造式の特徴を表現する空間(CDDD)に一度埋め込んだあと、線形表記(Canonical SMILES)として取り出す、という2段階の構成でした。またCDDDはオートエンコーダーによるものでした。
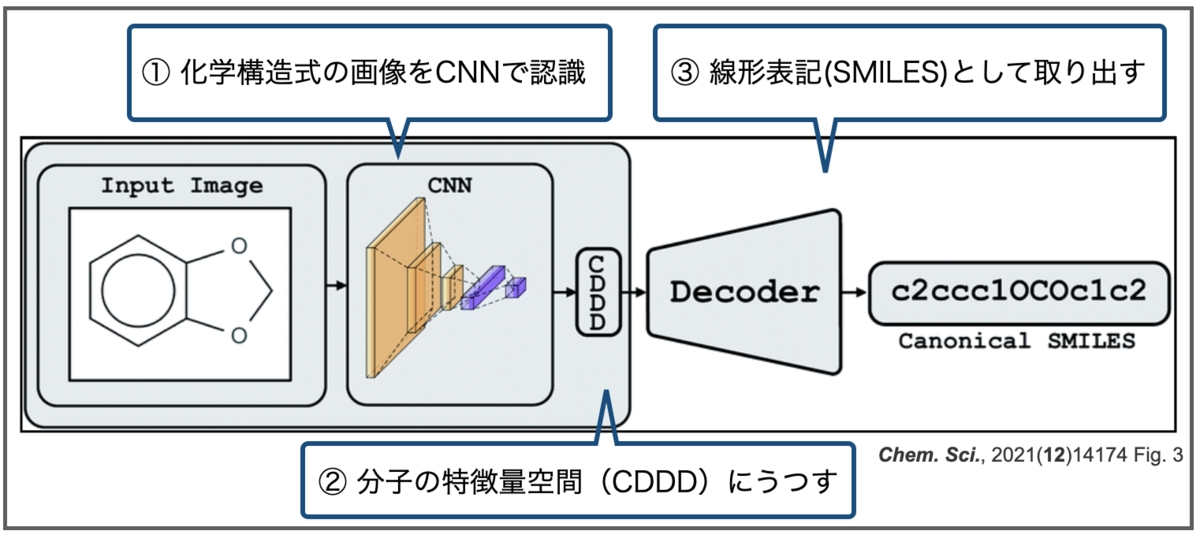
対してDECIMER 1.0も、画像を認識するための入力ネットワークと、認識した情報から化学構造式を取り出すためのネットワークの2段階の構成となっています。
ですが、化学構造を取り扱うネットワークを構築するために線形表記 SELFIESも利用していること、またTransformerベースのモデルになっていること、といった異なる点もあるようです。
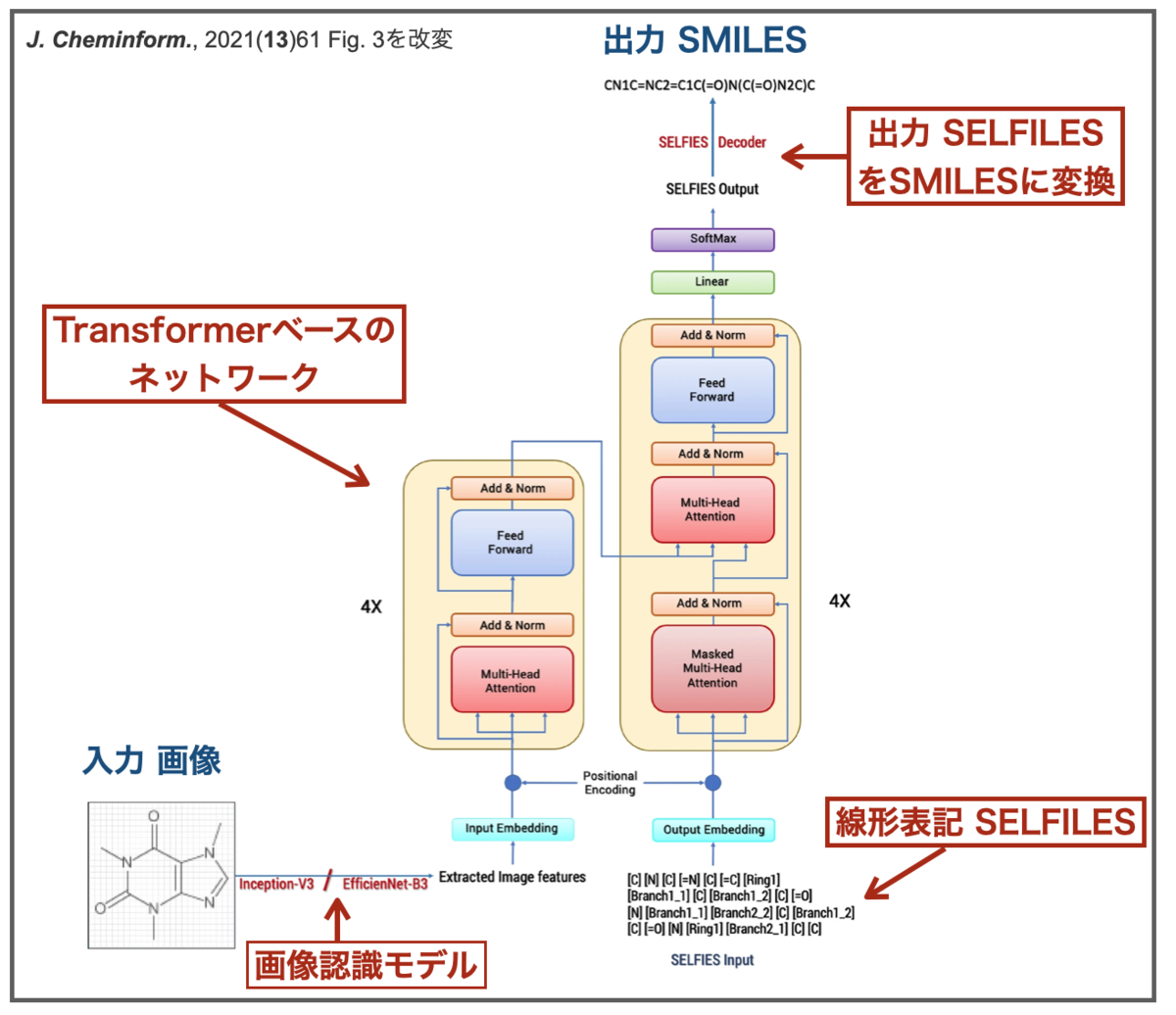
SELFIES (Self-referencing embedded strings)はAspuru-Guzik教授らのチームが開発した新しい化合物の線形表記方法で、この文法に従えばどんなふうに書いても100%分子として意味をなすという特徴があるそうです。
以下の記事が日本語でわかりやすく解説してくださっているのでおすすめです。
また、モデルのベースとなっているTransformerですが、こちらはAlphaFold2のベースにもなっていたやつですね。自然言語処理の分野で高い性能を発揮したらしく、よく使われているらしいです。
なお、前報DECIER(無印)ではオートエンコーダーをベースとしたモデルが使われています。DECIMER 1.0でもencoder-decoder型のモデルとTransformer型モデルが比較されており、結果、後者の方が精度が良かったそうです。
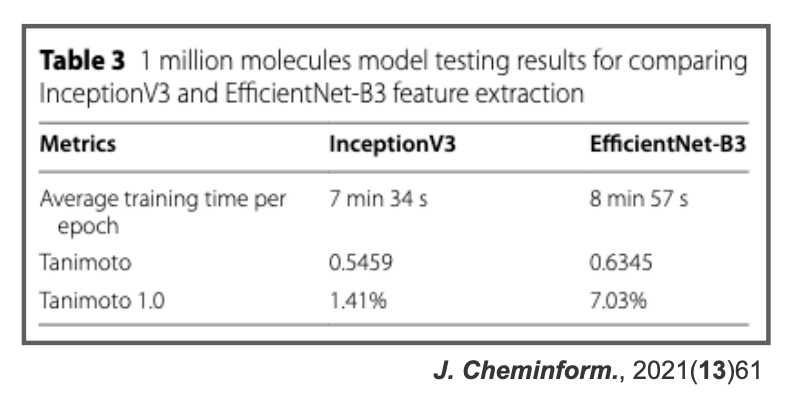
入力の画像認識 CNNのネットワーク構造としては、InceptionV3とEfficientNet-B3の2つを主に比較して、後者の方がパフォーマンスが良かった様です。
詳しくないのでよくわからないですが、各分野で最新の技術を色々と盛り込んだ感じでしょうか?最先端を攻める感じがアカデミックでいいですね!
1-3. DECIMER 1.0のデータセットと守備範囲
モデルの枠組みが大体わかったので、DECIMER 1.0の守備範囲についても参照しておきましょう!
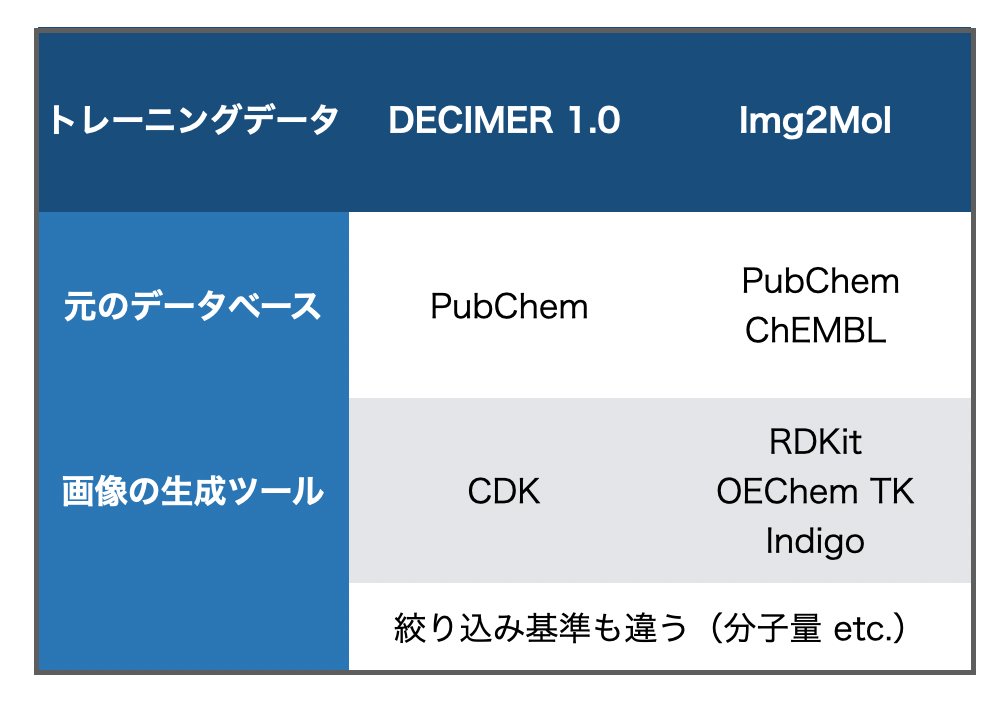
前回の記事では触れませんでしたが、Img2MolはPubChemとChEMBLから抽出し、一定の基準で絞り込んだ化合物をトレーニングに利用しています。また、トレーニング用の画像は複数のツール(RDKit、OpenEyeのOEChem TK、Indigo)を使って生成しています。
一方DECIMER 1.0は、対象データベースはPubChemのみ、トレーニング画像の生成はCDKのみを使用するといった、トレーニングデータセット準備段階での違いがあります。*2
個人的に面白かった点は、DECIMER 1.0は「立体化学やイオン化状態を含めるとどうなるのかか?」という検証にも挑戦しているところです。
論文中で使われているデータセットは以下の様に「立体化学/イオン化の情報があるか?」、「Image augmentationをしたか?」で3つに分かれています。
以下のTable 8がDataset 1に対する結果、Table 12がDataset 2に対する結果です。データセットのサイズが近いsubset同士を比較すると、Table 12の方がパフォーマンス(Tanimoto、Tanimoto 1.0)が低下する傾向にあるようです。
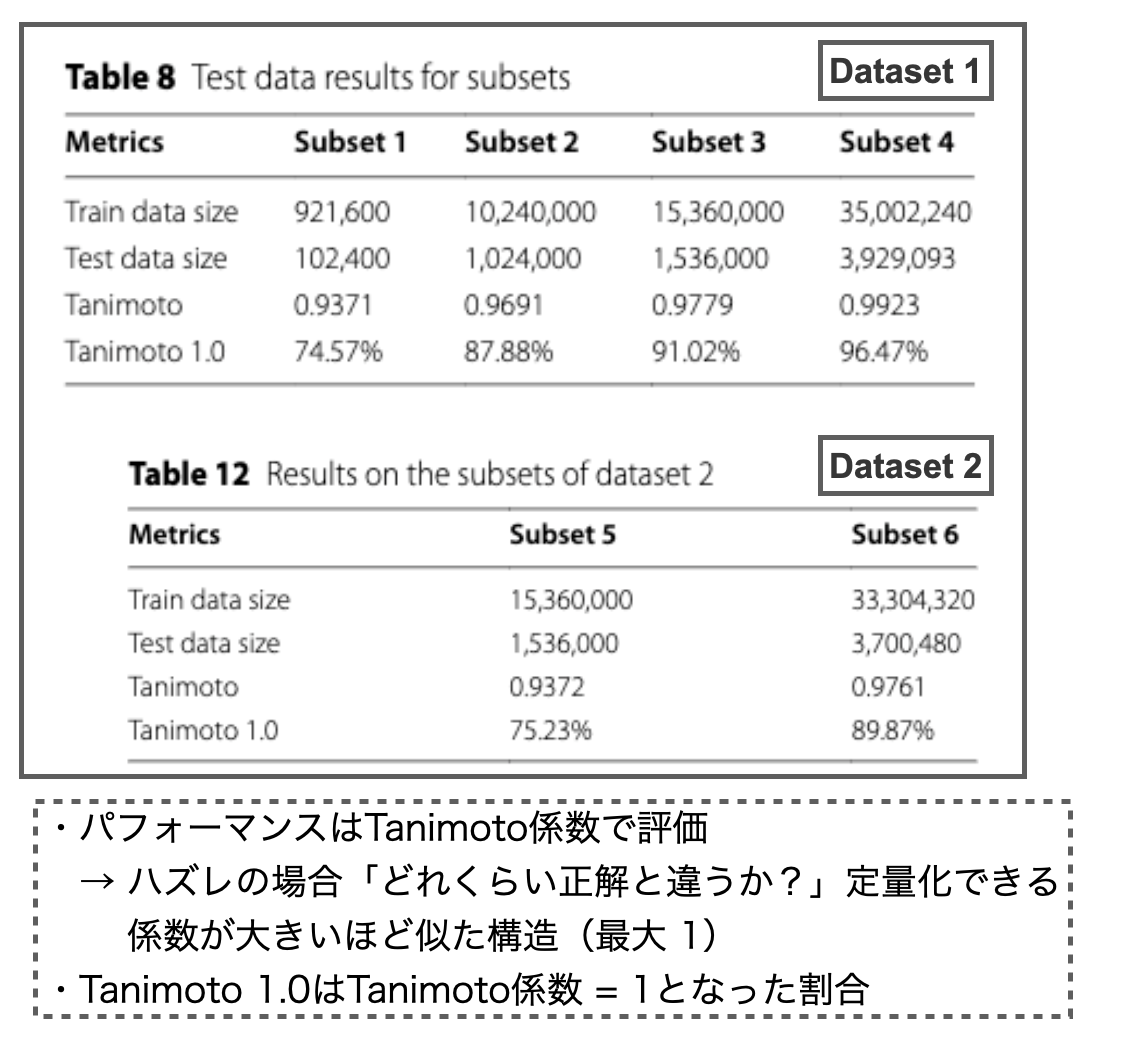
立体化学やイオン化状態の識別も考え始めると、問題が一気に複雑化するのでパフォーマンスが落ちるのは納得です。それでも結構良い結果が得られているように思いますが、皆さんはどう思われますか?
なおImage augmentationは、データ拡張を行ってDataset 2のデータを追加する(Dataset 3)とパフォーマンスが良くなるかを検証したものです。興味のある方は論文のFig. 10やTable 15あたりをご参照ください。
2. DECIMER 1.0で遊ぼう!
DECIMER 1.0の概要とImg2Molの違いがだいたい分かってきたので早速遊んでみましょう!
DECIMER 1.0はMITライセンスで公開されているので企業の方にも手を伸ばしやすいかもしれません。
後ほど記載しますが、学習済みパラメータはGoogle cloudのStorageにあり、プログラムを初めて利用するときにダウンロードする仕組みとなっているみたいです。
2-1. インストール
インストール方法はGitHubのREADMEの通りです。conda環境での利用が簡単なのでオススメとのことです。
# pipでgithubからインストール $ pip install git+https://github.com/Kohulan/DECIMER-Image_Transformer.git # もしくは以下でPyPiからインストールしてもOK $ pip install decimer
ところが、Img2Molの時と同様、Macユーザーはちょっとだけ面倒です。
MacOSでNvidia GPUがない場合は「tensorflow==2.3.0」を指定して入れなければいけないとの記載があります。上のコマンドでは私のMacにDECIMERをインストールできませんでした。
そこでgit cloneでDECIMERを落としてきた後、setup.cfgを少し書き換えるとインストールできました。
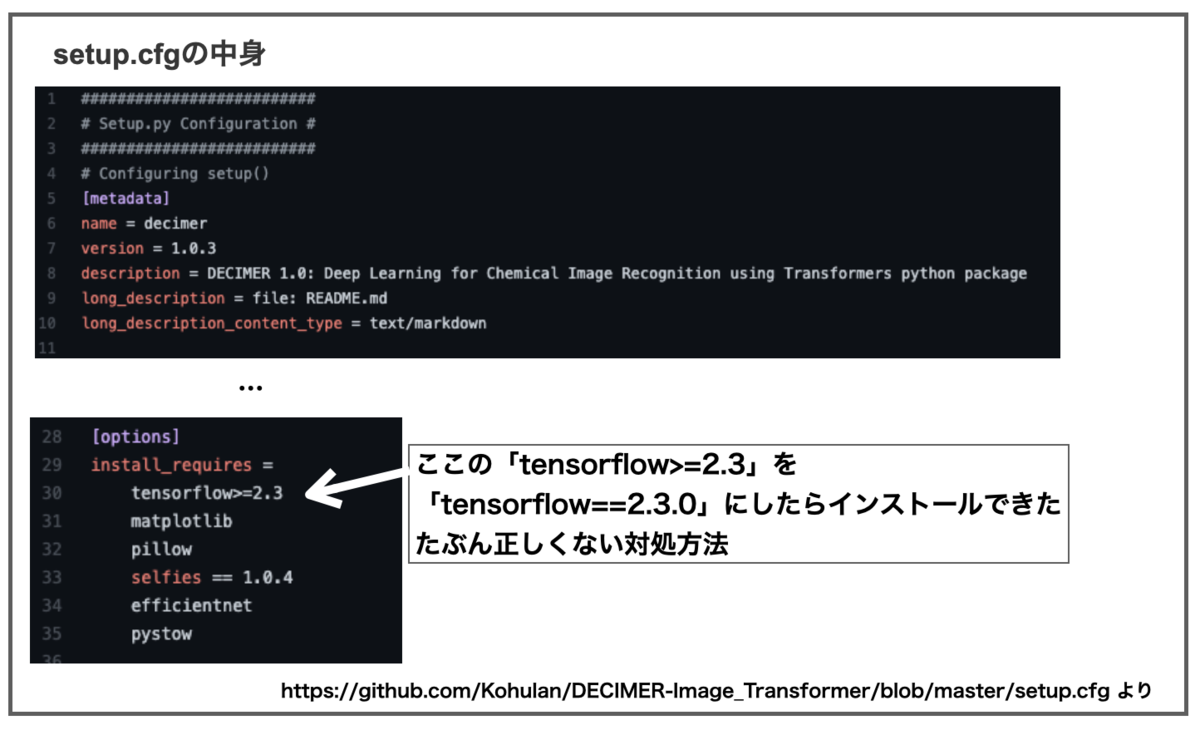
こういう感じの作業でした。
# githubからディレクトリを落としてくる $ git clone https://github.com/Kohulan/DECIMER-Image_Transformer.git decimer # setup.cfgを上図の様に書き換えて保存 # ディレクトリを移動 $ cd decimer/ # インストール開始 $ pip install -e.
どなたか正しい方法を教えてください。
2-2. 訓練済みパラメータの入手
先に書いた様に、DECIMER 1.0は学習済みパラメータも公開されていて、初回利用時にダウンロードされます。
私はパラメータの保存場所などが後でわからなくなるのを防ぐために、手動でダウンロードしてdecimerディレクトリの中に入れることにしました。
decimerのsrc/decimer/ディレクトリにあるDECIMER.pyをみると以下の記載があります。

デフォルトのディレクトリにパラメータファイルがなければmodel_urlからダウンロードしてくるみたいなことが書いてあります(たぶん)。
というわけでmolde_url = "https://~~~~" となっているURLにアクセスしてパラメータのzipファイルをダウンロードしました(1GBくらい)。
パラメータ保存場所として、DECIMERディレクトリにTrained_Modelsという新しいディレクトリをつくり、そこにzipを展開したファイルを格納しました。あとはDECIMER.pyやDECIMER_V1.0.pyの中にあるモデルのパスの箇所を書き換えて保存したら終わりです。
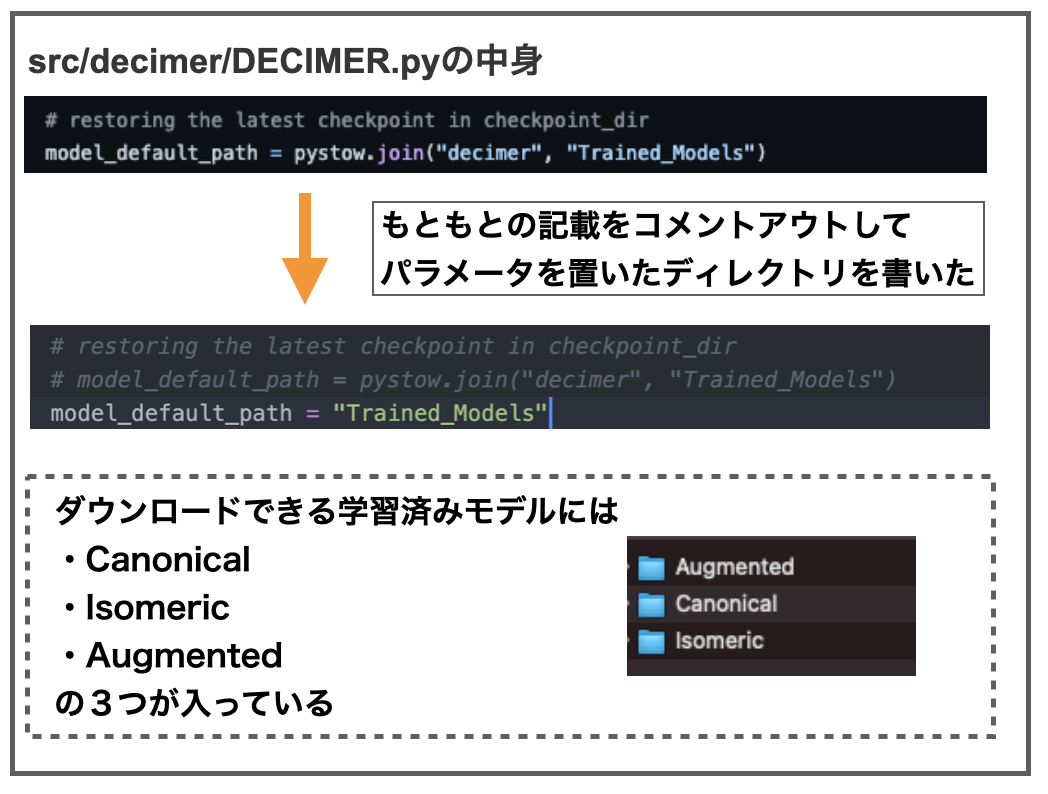
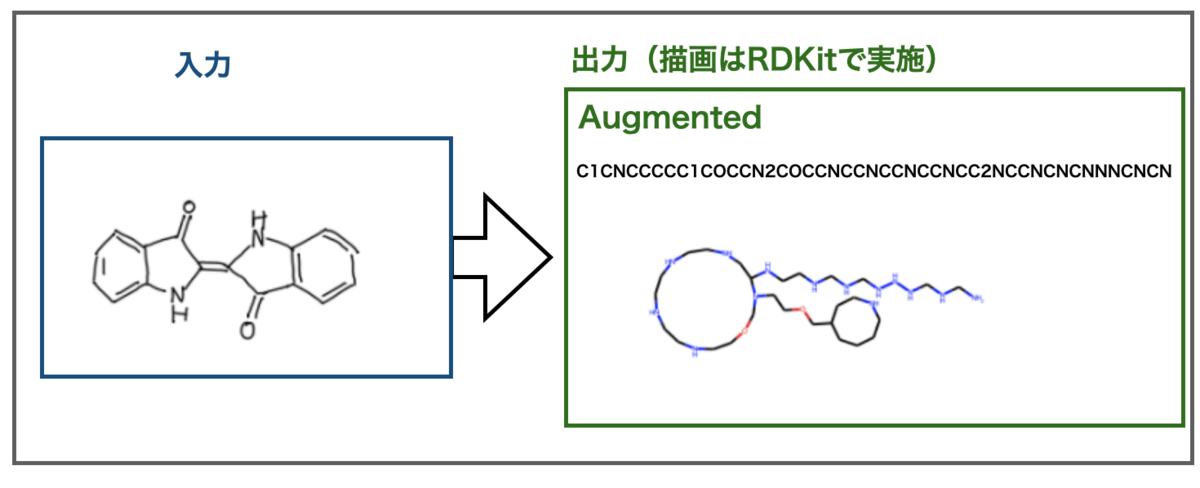
訓練済みのモデルにはCanonical、Isomeric、Augmentedの3つがあります。おそらく文献で触れられていた3つのデータセットに対応するものだと思います。*3
ダウンロードファイルの保存先を覚えておけばこんな余計なことをする必要はないと思います。私はすぐ忘れるんです。。。
何はともあれインストールとパラメータの準備ができました!
2-3. サンプルで遊んでみよう!
とりあえずJupyter notebookを起動して遊んでみましょう。Sample_Imagesディレクトリに画像例も準備してくださっているので利用します。
ライブラリをインポートしたあと、画像ファイルのパスと使いたいモデルを指定するだけでSMILESに変換して返してくれます。
from decimer import DECIMER # モデルの指定 model_name = "Isomeric" # 画像のパスを指定 img_path = "Sample_Images/caffeine.png" # 変換前に元々の画像を確認する from IPython.display import display from PIL import Image display(Image.open(img_path))
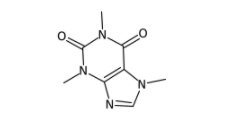
上のような画像を入力とします。さてDECIMERは無事SMILESに変換できるでしょうか??
# DECIMERでSMILESを予測 res = DECIMER.predict_SMILES(img_path, model_name) print(res) # > CN1C=NC2=C1C(=O)N(C)C(=O)N2C
SMILESができました!でもあってるのかわからない!読めない!
RDKitで変換した図をみてみます。(DECIMERと共存させられなかったので別のnotebookでやりました。)
from rdkit import Chem from rdkit.Chem import Draw m = Chem.MolFromSmiles("CN1C=NC2=C1C(=O)N(C)C(=O)N2C") Draw.MolToImage(m)
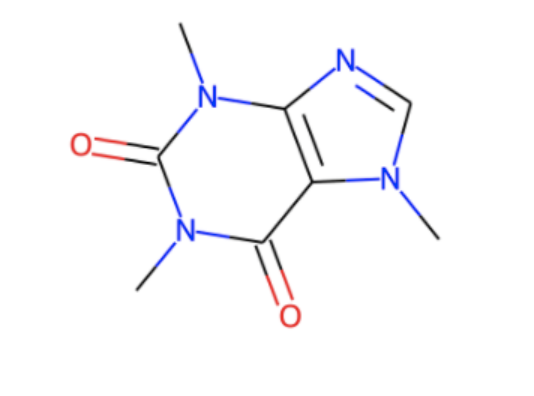
どうやらDECIMERはカフェインを認識してうまくSMILESに変換できたようです!
他のmodelではどうでしょうか?CanonicalとAugmentedで行った結果はこんな感じです。
AugmentedはIsomericと同様正しく認識できました!残念ながらCanonicalは全ての原子が硫黄の鎖状構造と誤認識してしまったようです。
論文を見る限りAugmentedが一番良さそうなので、以降の検証はこちらで行ってみます。
気になるのは立体化学の認識です。サンプルファイルに立体化学を含む例がありました。
どうだ!
おお!不斉点が認識されました!!SMILESに@が入っているだけでテンションが上がります!!
RDKitでの描画の都合上向きが変わってしまっていますが、立体化学も正しいようです。また、不斉の明記されていない炭素原子もありますが、ここも不斉情報がないものとして正しく認識できています。すごい!!
次に電荷を帯びたイオンはどうでしょうか??こんな例がサンプルにありました。
プラス電荷が認識されました!サンプルなのでうまくいくだろうとは思っていても実際に結果が出ると嬉しいですね!
話がそれますが、描画を並べてみるとRDKitの原子を色分けするスタイルはわかりやすくて良いですね。
2-4. 別のサンプルでテストしよう!
さてDECIMERの使い方がわかってきました。
次に気になるのはソフトの汎用性です。ソフトウェアのサンプル画像以外でもうまく機能するでしょうか???
比較対象としてちょうど良いのでImg2Molに提供されていたexamplesでテストしてみましょう!
カフェインに類似した構造の画像例です。
残念!だめでした。。。出力は芳香族がうまく認識できておらず、窒素-硫黄結合といった構造もできてしまっています。
別の例ではどうでしょうか?
こちらもだめでした。今回は芳香環を一つ認識できたましたが、なんとなく似ていそうで全然違う結果になってしまいました。
ひょっとして画像の化学構造式に色が含まれているのが悪かったりするのでしょうか?
別の画像例としてWikipediaのプロリンを使ってみましょう。元素記号が大きく、原子の色分けがない画像です。こちらもImg2Molではうまくいったものです。
なんだかすごい構造になりました。。。こんなアミノ酸は嫌だ。
最後に手書き画像の例を見てみましょう。Img2Molのサンプルです。
こんな結果になりました。
DECIMER 1.0の文献中にも記載がありましたが、環構造の認識に失敗すると、ヒトの目にはとても大きな間違いにみえますね。
上手くはいきませんでしたが手書きの構造式も化学構造として認識できいるという点はなかなかすごいと思います。
3. おわりに
以上、今回は化学構造式のOCRソフト DECIMER 1.0で遊んでみました。Img2Molと比較していかがだったでしょうか?
個人的には立体化学の認識、電荷の認識にも挑戦していて、サンプル画像ではうまく認識できているという点が驚きでした。Img2Molは立体化学を全て無視するようでしたのでDECIMERの長所だと思いました。
一方で、サンプル画像以外の画像では認識があまり上手くいっていないという印象でした。Img2Molは小さな分子であればWikipediaやPubChemのキャプチャ画像でも認識できたので、汎化性能(?)という点では課題があるかもしれません。
このあたりトレーニングデータセットの作り方による差もありそうです。Img2Molでは複数のツールで入力画像を作っていたのに対し、DECIMERはCDKのみでした。それぞれ異なる描画のクセがあるソフトを組みあせたデータセットの方がバラエティがあり、汎化性能が高くなりそうです。
DECIMER 1.0をImg2Molのデータセットでトレーニングしたらどうなるか?気になるところです。
あくまで素人の感想です。すみません。。。
また、DECIMERプロジェクトはImg2Molで今後の課題となっていたセグメンテーションにも既にWebアプリを公開しています。OCSRに関わる複数のソフト群全体の開発として取り組まれているので、シナジー効果によりさらに精度が改善していくかもしれません。次報が楽しみですね!
今回も色々と間違いが多そうです。ご指摘いただけると嬉しいです。
ではでは!
化学構造式のOCRソフト Img2Molで画像をSMILESに変換しよう!
少し前になりますがKaggleでBristol-Myers Squibb - Molecular Tranlationというコンペティションが開催されていました。
化学構造式を含む画像をその化学構造のInchi(International Chemical Idendifier)文字列に変換する精度を競うものでした。*1
巨大製薬企業のブリストル・マイヤーズスクイブがこのようなコンペティションを行うくらいなので、化学構造式のOCR(Optical Charactier Recognition)、「画像から化学構造式情報をコンピュータ処理できる形で抜き出すタスク」がニーズがありかつ難しいものであることが分かります。
さて今年のRDKit UGM 2021でも同様のタスクについての発表がありました。
その名もImg2Mol‼︎
- 発表の動画: YouTube : Djork-Arné Clevert: Img2Mol – SMILES Recognition from Depictions ofChemical Structure
- 発表のスライド:Clevert_Img2Mol.pdf
- 文献 : Img2Mol – accurate SMILES recognition from molecular graphical depictions Chem. Sci., 2021,12, 14174-14181
- GitHub : bayer-science-for-a-better-life/Img2Mol
そんな難しいタスクのためのプログラムが公開されている??
それなら遊ぶしかない!やってみましょう!
文献を流し読み
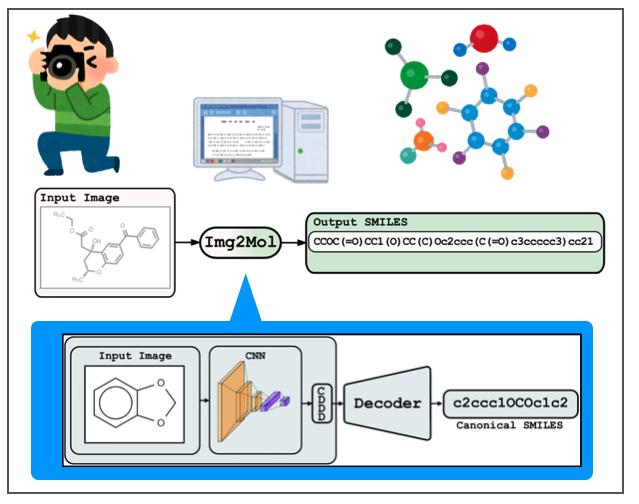
Img2Molはバイエルの機械学習チームが開発したプログラムで、化学構造式の画像をSMILESに変換してくれます。文献がオープンアクセスになっているのでざっと眺めてみます*2。
化学構造式OCRのモチベーション
そもそもなぜ化学構造式を画像から機械処理できる形で取り出すタスクに需要があるのでしょうか?
それは創薬化学の重要な情報源である文献と特許がともにコンピューター処理に適した形で情報を提供していないからです。
最近ではJournal of Medicinal ChemistryなどSupplementary Informationで化学構造のSMILESを列挙したファイルを提供するものも出てきましたが、まだまだ多くの文献で化学構造式は画像でのみ示されています。また特許では公開されている書類がPDFや画像形式であり、さらに産業上の理由から敢えて曖昧に書かれていることもあります。
このような状況なので、最新の情報を効率的に2次利用できる形でとりだしアップデートしていくのはとても大変です。というわけで画像から構造式をとりだすことにニーズがあるそうです。
ところで生理活性物質の公開データベースとしてみんな大好きなChEMBLも、専門家による手作業のキュレーションで情報更新されているようです。維持・運用のコストを考えると感謝しながら使わないといけませんね。
Img2Molのアプローチ
だいたいモチベーションがわかりました。では実際にタスクをこなすにはどうすれば良いでしょうか?
課題としてはこんな感じです。
1. 画像中の構造式を認識する
2. 認識した構造式をコンピューター処理できる形に取り出す
3. 取り出した構造は化学的に妥当なものである必要がある
1.と2.は全てのOCRに共通で、3.は化学ならではの課題です。
で、この化学ならではの部分をどうするか?
Img2Molでは画像を畳み込みニューラルネットワーク(CNN)で認識し、化学構造式の特徴を表現する空間(CDDD)に一度埋め込んだあと、線形表記(Canonical SMILES)として取り出す、という2段階の構成をとっているようです。

CDDDはcontinuous and data-driven molecular descriptorsの略で、同じくバイエルのチームが開発した記述子です。こちらの論文もOpen Accessになっています。
- Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations Chem. Sci., 2019,10, 1692-1701
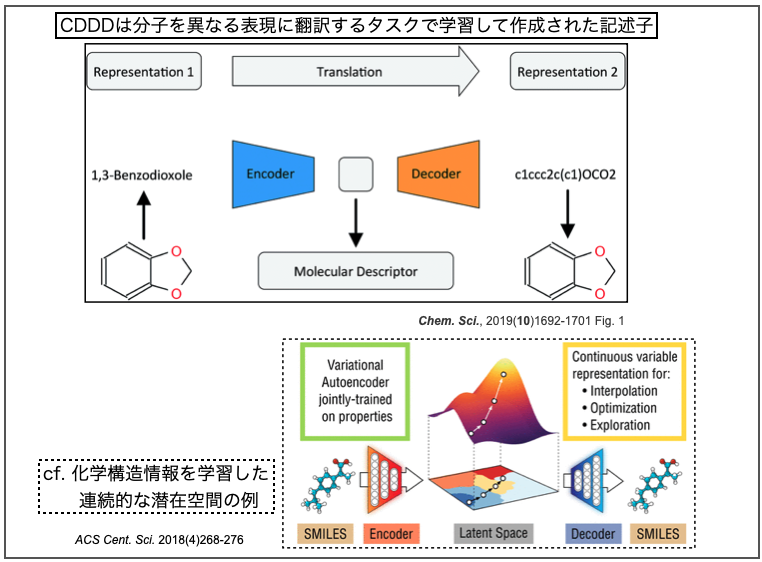
オートエンコーダ(Autoencoder、自己符号化器)はニューラルネットワークの一つで、「入力されたデータを一度圧縮し、重要な特徴量だけを残した後、再度もとの次元に復元処理をするアルゴリズム」だそうです*3。
ある化学構造を表すSMILESには複数通りの書き方があり、それらを入力として(一意の)canonical SMILESを出力する翻訳のタスクをこなすオートエンコーダを学習します。このネットワークは化学構造の特徴を学習した潜在空間(latent space)となっています。なので、化学構造を表す記述子として使うことができ、異なるタスクにも色々と応用できるよ!(とかそんな話です。たぶん。)
上図で引用した下半分はAspuru-Guzik先生のチームによる以下の論文です。*4
- Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules ACS Cent. Sci. 2018, 4, 2, 268-276
ところで、バイエルのチームはImg2Molと同様のアプローチを以前にも報告しています。以下の論文では「ECFP記述子を入力として、もともとの分子構造を逆に推定するリバースエンジニアリング」を行っています。
- Neuraldecipher – reverse-engineering extended-connectivity fingerprints (ECFPs) to their molecular structures Chem. Sci., 2020,11, 10378-10389
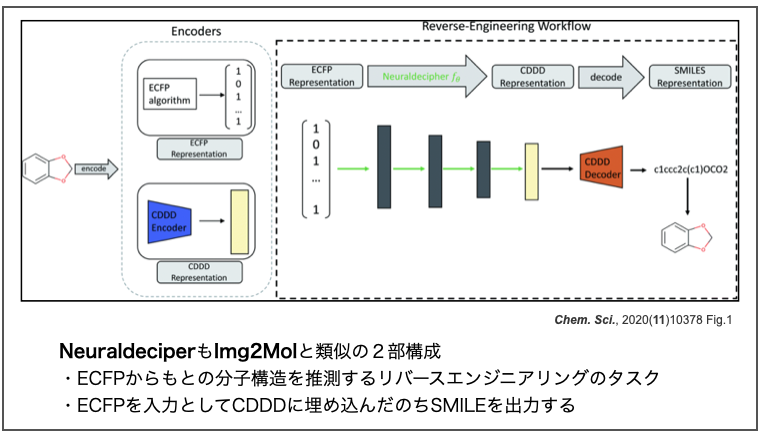
このタスクでも一度CDDDに埋め込んだ後SMILESとして出力しています。応用範囲が広い記述子を開発できると便利だなー。
Img2Molのパフォーマンス
Img2Molのネットワーク構造やメソッドの詳細はよくわからないので割愛します。ごめんなさい。
結果っぽいのだけ貼っておきます。
他の手法よりパフォーマンスが良かったよの図(ベンチマーク)。
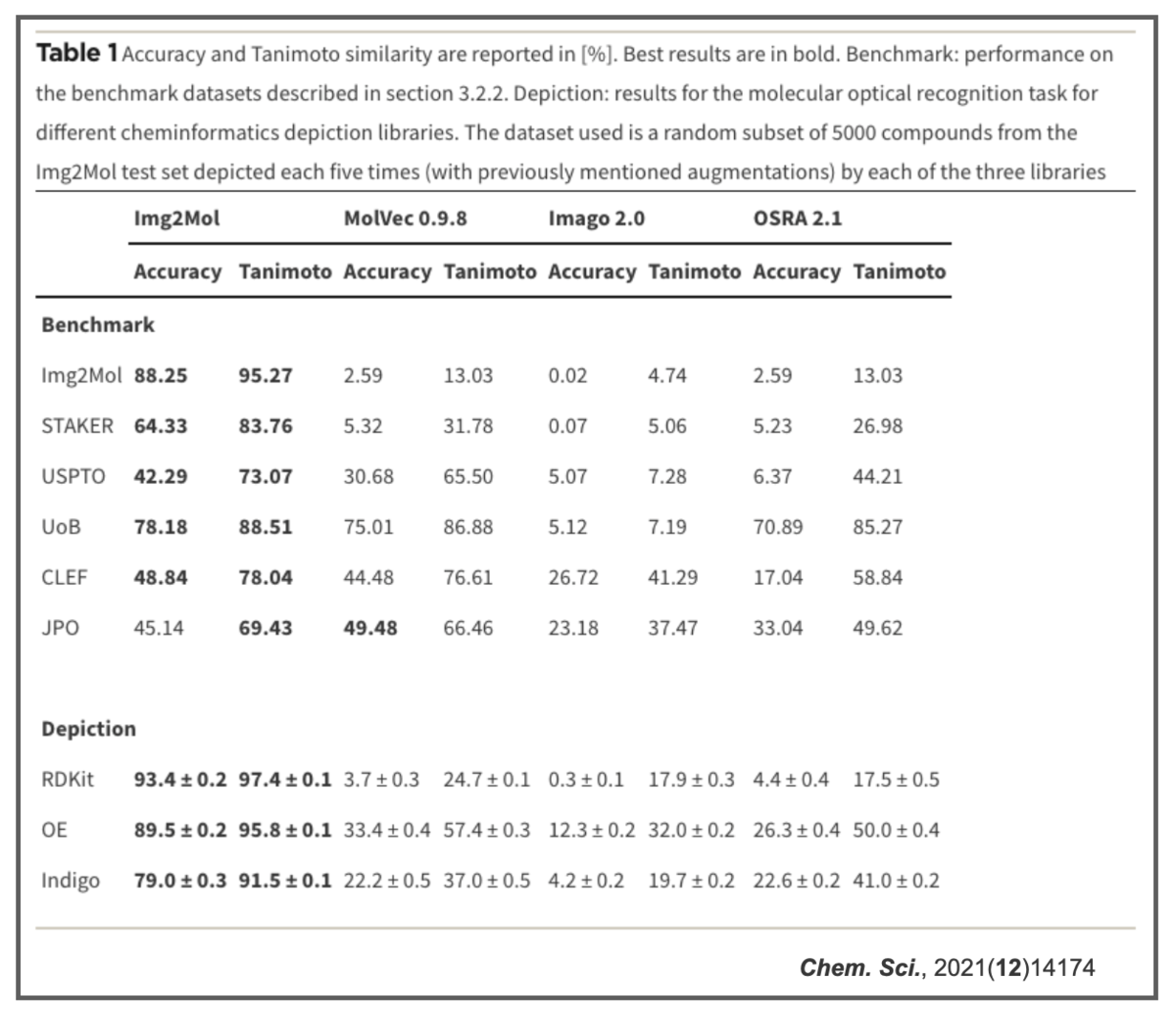
AccuracyだけでなくTanimoto類似度でも評価しているのが面白いです。
学習に使う化学構造を描くツール(depiction)によっても結果が変わっているのも興味深いです。同じ化学構造でもソフトによって表現・相性が変わるところに、コンピュータで化学を扱う難しさを感じます。
さらに面白い結果はこちら。画像の解像度(Resolution)とパフォーマンスの関係です。なんとImg2Mol (no aug)は入力の解像度が上がると精度が落ちています。
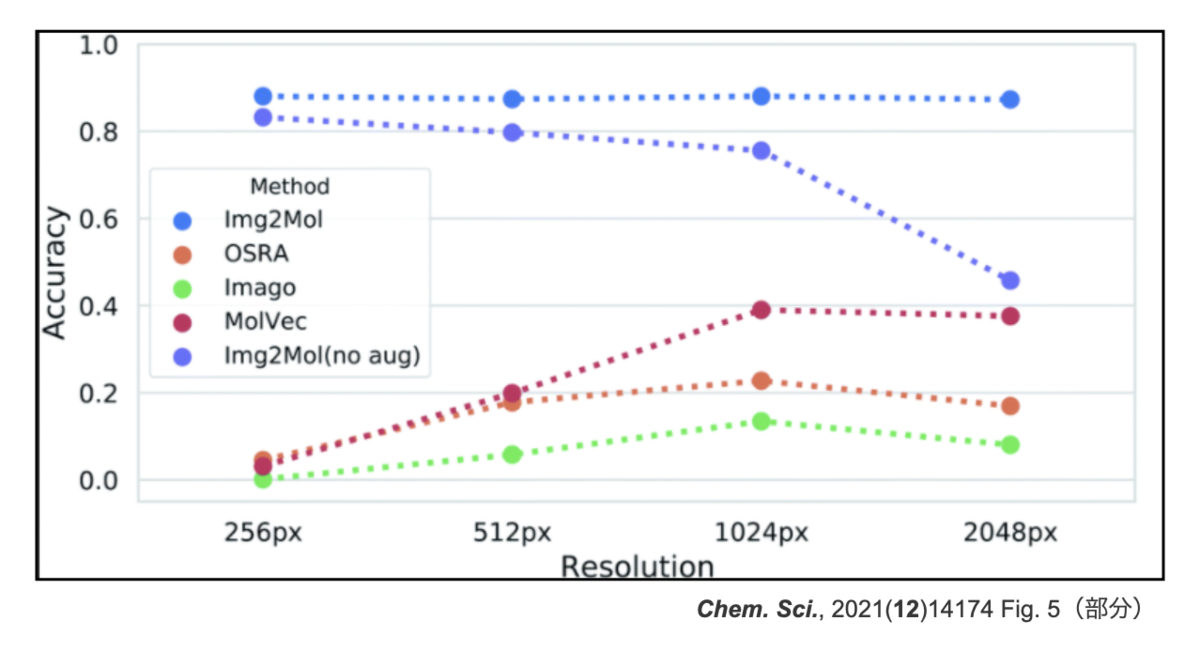
「入力の質が上がって精度が悪くなるの??逆じゃないの??」となりますが、どうやらネットワークへの入力の仕方によるもののようです。
ネットワークの構造は決まっているので、入力画像は全て同じ形(224 x 224 px)にリサイズされます。高解像度の画像は、このリサイズの処理で失われる情報が逆に多くなるため、ぼんやりとした入力になってしまうようです。
この解像度の問題や分子サイズの大きな構造の認識が難しいという問題を解決するため、data augmentationをおこなって学習し直しています。これで得られたよりロバストなネットワークが上図の「Img2Mol(無印)」に相当するものです。高解像度でもパフォーマンスが落ちていません。
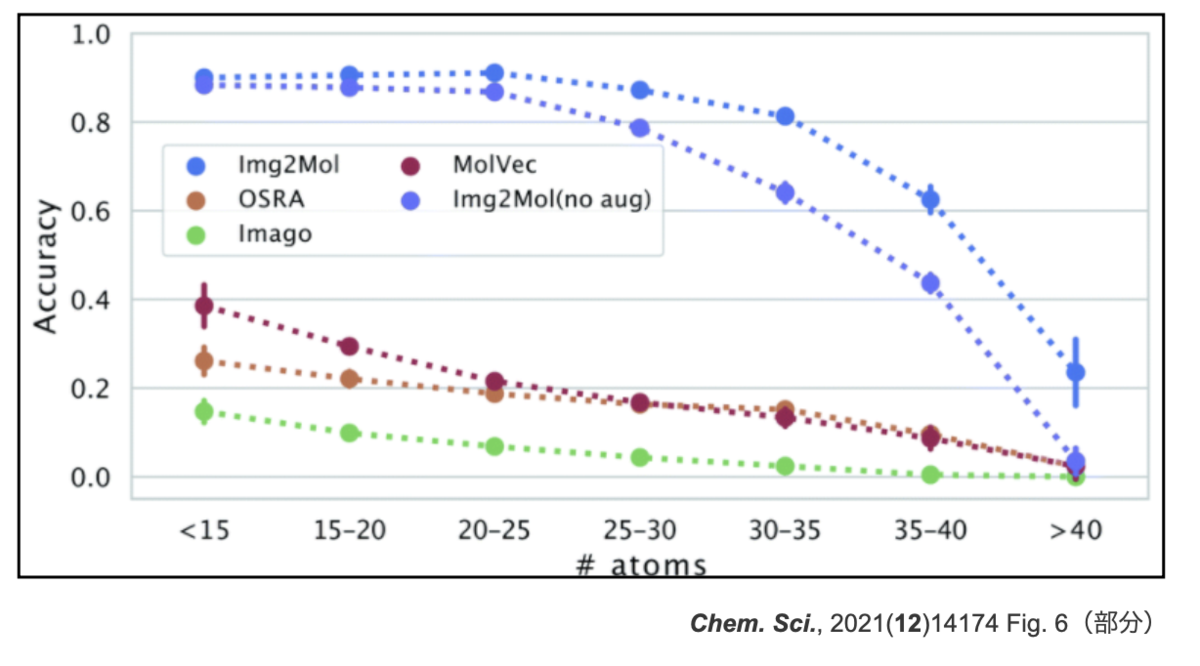
分子サイズとパフォーマンスとの関係も貼っておきます。こちらもaugmentationで改善はしていますが、原子数が30を超えたあたりから精度が落ちてきています。
以上、Img2Molのざっとした文献紹介でした。正確な情報は論文やRDKit UGMの発表動画などをご参照ください。
Img2Molで遊ぼう
それでは早速、Img2Molで遊んでみましょう!素晴らしいことにGitHubで公開してくださっています!
但し、学習済みパラメータのライセンスはCC BY-NC 4.0でnon-commercial use onlyですので商用利用できないことには十分ご注意ください(念のため)。*5
インストール
インストール方法はcondaでOKで簡単です。GitHubのREADMEの以下をコピペするだけ。
git clone git@github.com:bayer-science-for-a-better-life/Img2Mol.git cd Img2Mol conda env create -f environment.yml conda activate img2mol pip install .
・・・が、Macユーザーの方はご注意ください。Img2Molに必要なソフトとして「cudatoolkit=11.0」の指定がありますが、NVIDAがmacOSのサポートを終了したらしく、「CUDA ToolKit v11」はmac版がありません。*6
Img2Molのインストールに使う「environment.yml」ファイルは以下のようになっています。
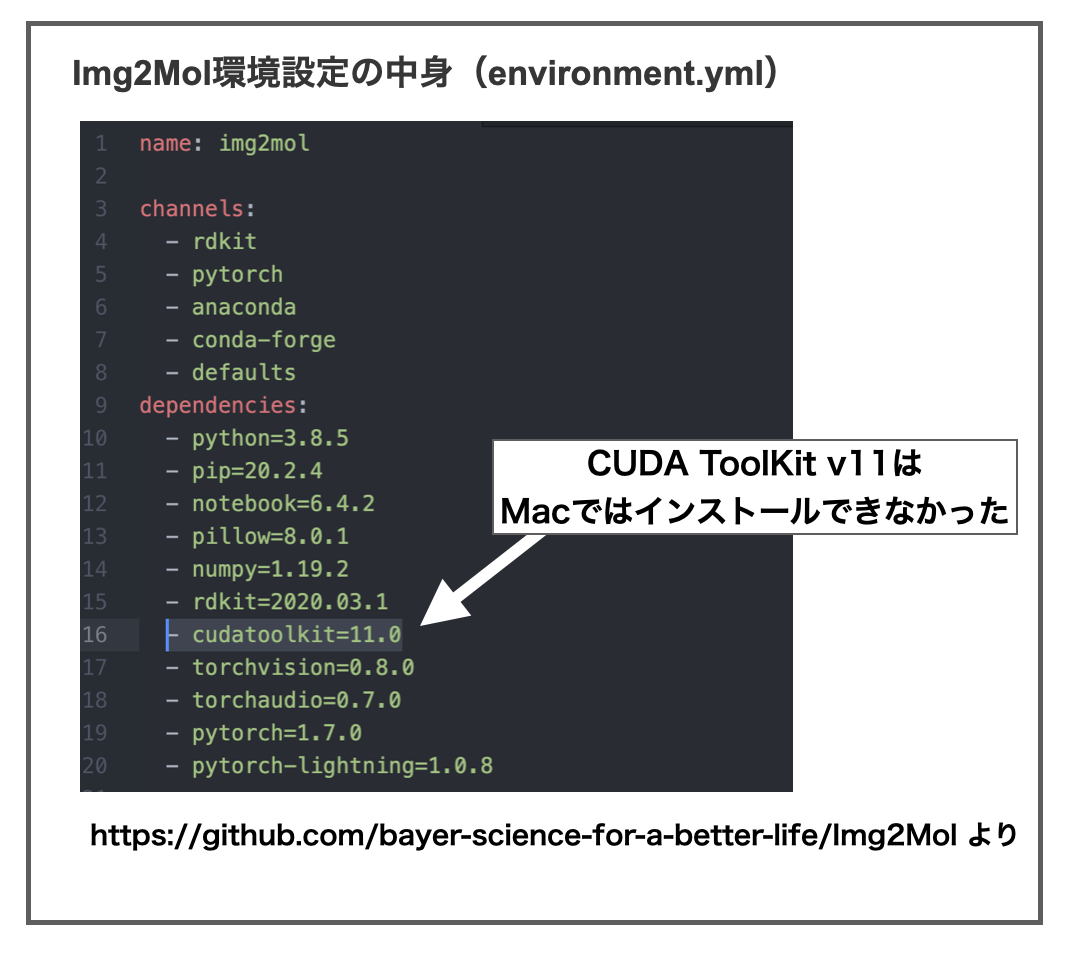
このままだと「condaチャンネルにそんなの無いよ!」って言われます。なので「cudatoolkit=11.0」のバージョン指定「=11.0」を消して「cudatoolkit」だけにして上書き保存してから再実行するとインストールできました。
旧バージョンのCUDA ToolKitが入るので動作の保証はできませんが、とりあえず動いたのでヨシ!*7
学習済みパラメータファイルはGoogleDriveからダウンロードできるようにしてくださっています(→ https://drive.google.com/file/d/1pk21r4Zzb9ZJkszJwP9SObTlfTaRMMtF/view )。大きい(~2.4GB)ので空き容量とインターネット回線にご留意を!
「model.ckpt」というファイルがダウンロードされるので、Img2Molディレクトリの中の「model」ディレクトリの中に放り込めば準備完了です。
Exampleで遊ぼう
準備ができたらJupyter notebookを開いて遊びましょう!
使い方がexample_inference.ipynbに、画像例がexamplesディレクトリにあるのでそのまま使わせていただきます。
# 必要なライブラリのインポート import torch from img2mol.inference import * from IPython.display import display # img2molを使う準備 device = "cuda:0" if torch.cuda.is_available() else "cpu" img2mol = Img2MolInference(model_ckpt="model/model.ckpt", # パラメーターファイルの指定 device=device) cddd_server = CDDDRequest() # > Initializing Img2Mol Model with random weights. # > Loading checkpoint: model/model.ckpt # > Setting to `self.eval()`-mode. # > Sending model to `cpu` device. # > Succesfully created Img2Mol Inference class.
これでImg2Molを使って画像からSMILESを予測するためのクラスが作成できました!
あとは変換したい画像のファイルパスを指定すれば予測を実行できます。
# exaplesディレクトリのdigital_example1.pngファイルを入力 res = img2mol(filepath="examples/digital_example1.png", cddd_server=cddd_server)
結果は辞書型で以下のようになっています。
print(type(res)) # > <class 'dict'> res # > {'filepath': 'examples/digital_example1.png', # > 'cddd': None, # > 'smiles': 'Cn1c(=O)c2c(nc(Sc3ccccc3)n2C)n(C)c1=O', # > 'mol': <rdkit.Chem.rdchem.Mol at 0x7fb09587eda0>}
入力ファイルとCDDD、結果のSMILESとRDKit Molオブジェクトも入っています。
今回入力に使ったのは以下の画像。
input_img = Image.open(res["filepath"], "r") display(input_img)
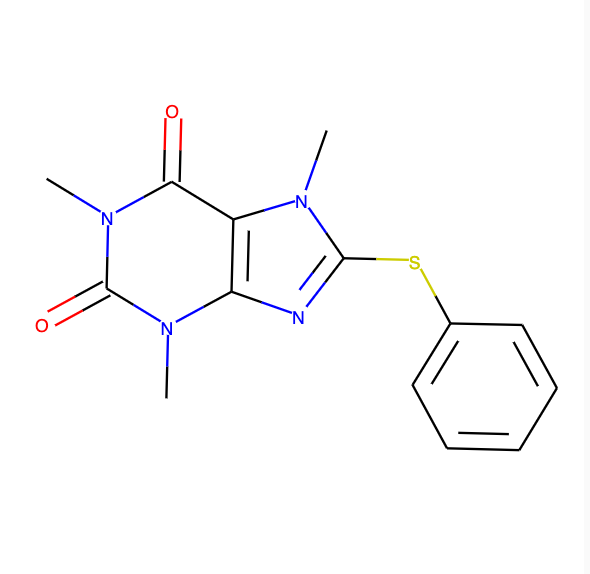
予測結果はこんな感じ。RDKit Molオブジェクトなのでjupyter notebookで構造式をそのまま描画できます。
# 変換後のSMILESを表示 print(res["smiles"]) # > Cn1c(=O)c2c(nc(Sc3ccccc3)n2C)n(C)c1=O # RDKit Molオブジェクトの構造式を描画 res["mol"]
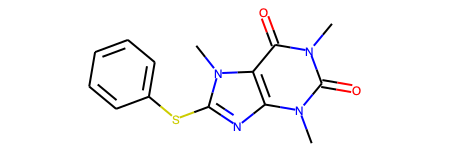
おお!正しく認識できてます!
私はSMILESを読めないのでわかりませんが構造式で確認できるので便利!!
手書きの構造式の例も用意されています。こういうのです。
# handwritten_example2.pngファイルを入力 res = img2mol(filepath="examples/handwritten_example2.jpg", cddd_server=cddd_server) input_img = Image.open(res["filepath"], "r") # 入力画像の確認 display(input_img)
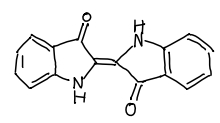
結果はこんな感じ
# 変換後のSMILESを表示 print(res["smiles"]) # > O=C1C(=C2Nc3ccccc3C2=O)Nc2ccccc21 # RDKit Molオブジェクトの構造式を描画 res["mol"]
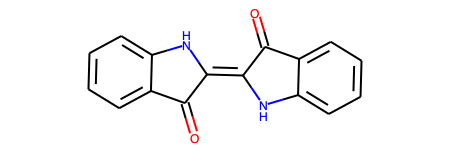
こっちもうまく認識できています!手書きの認識ができるなら授業のメモの電子化にも使えそうです!すごい!!!
ちなみに失敗例も提供されてたりします。コードは同じなので省略します。
こんなのです。
真ん中の環構造がフェニルではなくインドールとして認識されています。入力画像の右側ヘテロ環の結合関係がうまく認識できていないようです。
手書きの構造式は結合の長さや角度がまちまちだったり、元素記号のアルファベットが崩れていたりと、色々難しいポイントが多そうです。
いろいろな構造で遊ぼう
使い方が分かったので色々な画像を認識させてみましょう!
今年も色々な分子が話題となりましたが、なんといってもノーベル化学賞が有機触媒の開発に贈られたことは一大ニュースでしたよね!
というわけでプロリン画像をWikipediaから持ってきました(Wikipedia-プロリン)。こんな結果です。
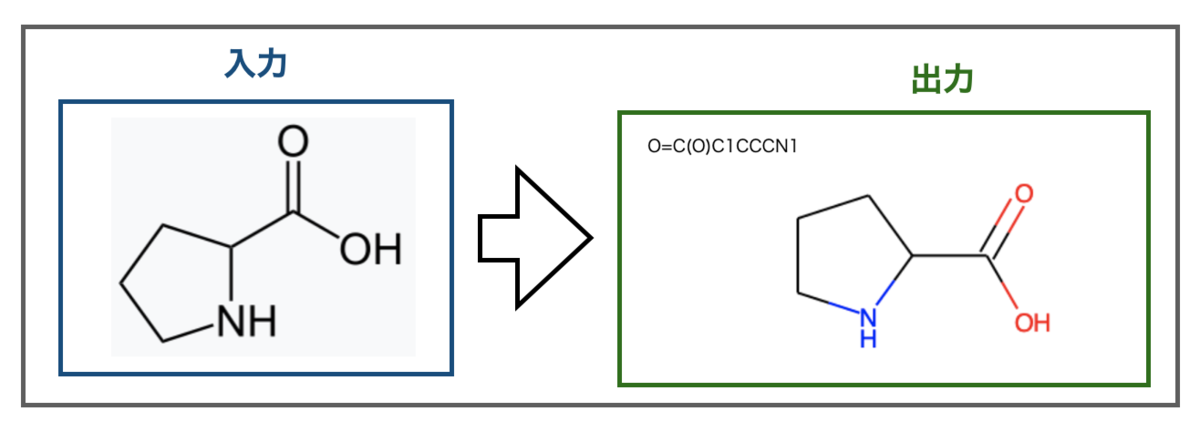
うまくできました!
マクミラン触媒はどうでしょうか?PubChemの構造を使ってみます((5S)-5-Benzyl-2,2,3-trimethylimidazolidin-4-one)。こんな感じです。
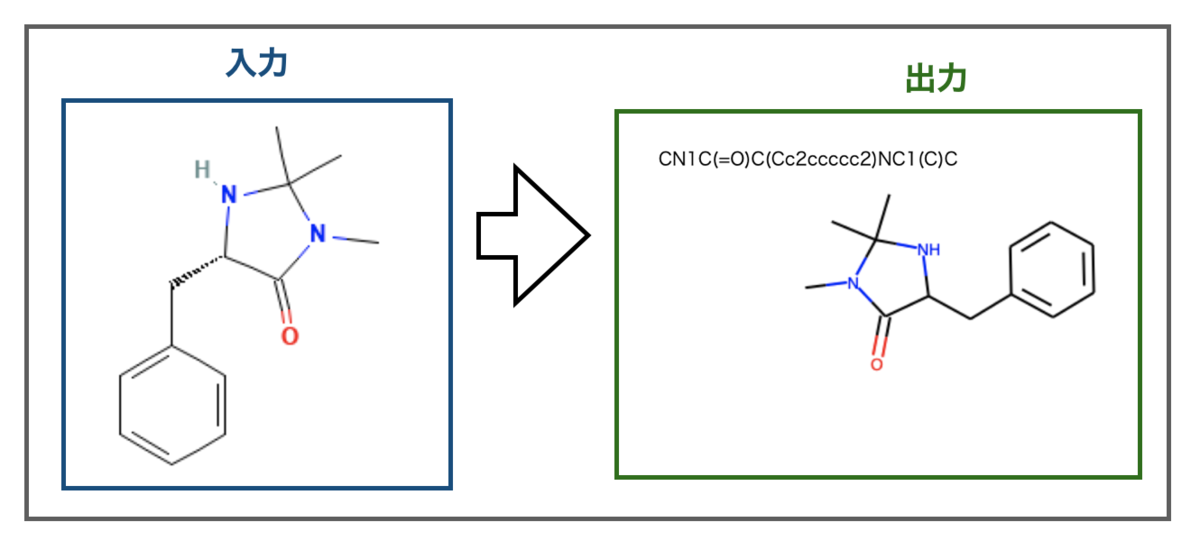
上手くいきました・・・が、不斉の情報までは認識できていないようです。出力のSMILESにキラル情報(@文字)がなく、構造式も不斉がありません。
ぱっとみた感じCDDDの学習で不斉の取り扱いまでは行っていないようなので、不斉情報は記述子に埋め込まれていないのかもしれません。
ところで化学構造の描画方法には今まで見てきたもの以外にも色々あります。Ball and StickやSpace-Fillingといったモデルで化学構造を表した画像はどのように認識されるのでしょうか?
同じくPubChemから画像を作成してImg2Molに投げてみました。
Ball and Stick描画したマクミラン触媒はこんな結果になりました。
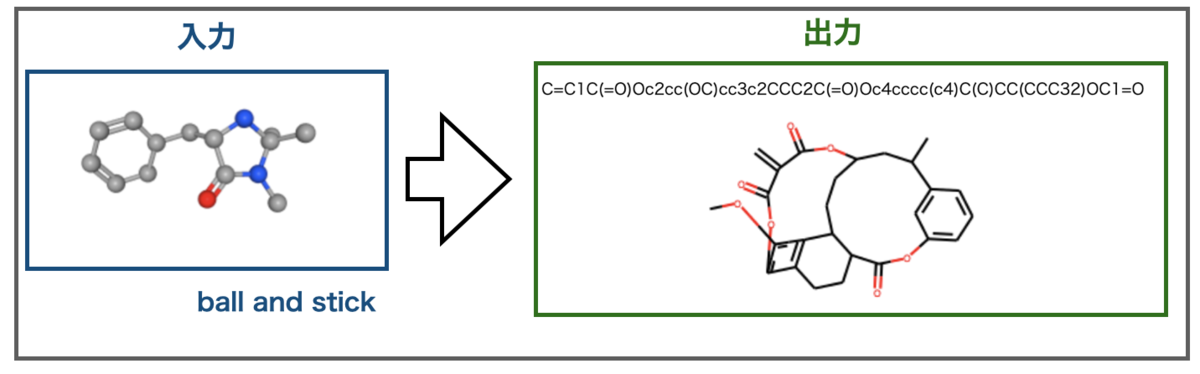
すごい構造。。。出力には窒素原子が含まれていませんが、色で原子を区別した描画では区別できないのでしょうか?
次にSpace-Fillingで描画したマクミラン触媒はこんな結果でした。
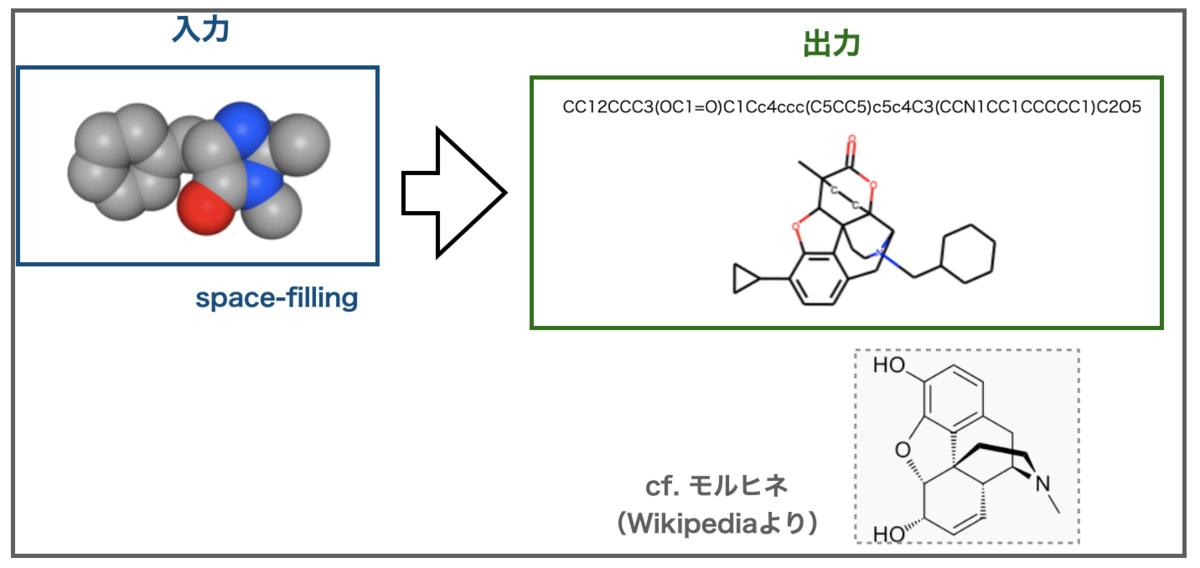
面白いことにSpace-Fillingの出力はモルヒネ様の骨格に認識されました。さらに今度は窒素原子も入っています???
出力に含まれる酸素原子の数を考えると、色で窒素を認識できていると思えないのでモルヒネ様に誤認識して引きづられた結果かもしれません。Img2Mol内部でどの様に認識されているのでしょうか???
不適切な入力構造はここまでにして。。。。
さて論文中では分子のサイズ(原子数)が大きくなると精度が落ちるとの記載がありました。最後に試してみましょう!
今年話題になった分子といえばKRAS G12C阻害剤として承認されたAmgenのSotorasibがありますね(Wikipedia - Sotorasib)。組成式C30H30F2N6O3なので、水素原子を除いても原子数41となります。
さてどうなるでしょうか?
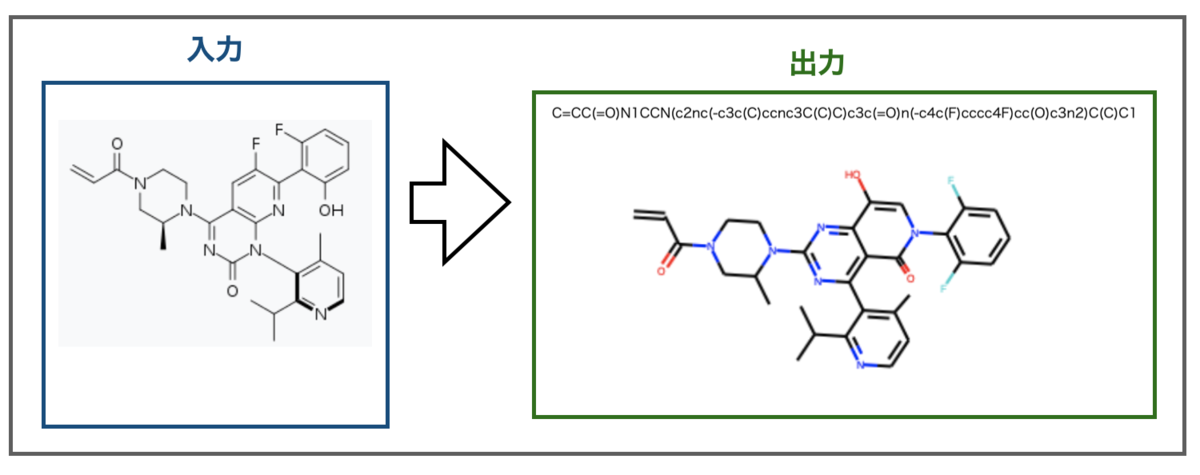
ぱっと見認識できている様にも見えますが、ちょっとずつ違います。
骨格の真ん中の環構造はカルボニルの位置や他の環との接続の仕方が異なります。また右端のフェニル基はオルト位に置換があるのは良いもののどちらもフッ素原子になってしまっています。入力画像の真ん中の環のフッ素原子と位置が近いので誤認識したのかもしれません。
画像からの構造の読み取りが失敗したとしても大枠が正しいなら細かい修正だけで済むので良いかな?とも思いましたが、微妙にちょっとずつ違うと間違い探しみたいで逆に難しくなってしまうかもしれません。
課題
いくつか遊んでみた結果、かなり精度が高く便利なツールという印象ですがいくつかできないこともわかってきました。
- 不斉の認識
- 分子サイズの大きなもの
また、利用を想定されていないであろう描画方法(ex. Ball and Stick, Space-Filling) 、色で表された原子の識別はできませんでした。
化学構造の特徴を学習した潜在空間にうつすならひょっとしていけるかも…と思いましたがダメでした。無茶を言ってすみません。
この他にRDKit UGMの動画で指摘されていたものにはキャプションつきの画像がありました。化学構造式以外の情報(化合物名など)が同じ画像の中にあると区別して認識できないとのこと。
「画像のセグメンテーションをどうするか?」が次の課題のようです。
おわりに
以上、今回は化学構造式のOCRソフト、Img2Molで遊んでみました。課題はあるもののかなり便利なツールだなぁと思いました。
ところで国語の成績も悪かった私ですがいまだに覚えている短歌があります。
「のみ込めぬままに図表は消されゆき 遠き席にて聴き終えて立つ」
詠み手*8が学生の時、講義の内容が理解できないうちに板書が消されてしまい、そのまま講義が終了してしまって「あーあ、やんなっちゃったよ」みたいな内容だった気がします。
なぜ覚えているか???
同じ経験をたくさんしたからです!
そう!化学構造式が超高速で現れては消えていく有機化学の授業でね!!
Img2Molがあればパシャーと写真撮って、後で画像からいい感じで構造式を抽出できたかもしれません。。。
今回も色々と間違いが多そうです。特に論文の中身。ご指摘いただけると嬉しいです。
おしまい。
*1:DeNAのデータサイエンティストの方々が参加されたチームが3位入賞・金メダルを獲得されたそうです(DeNA News)。格好良い!
*2:CC BY-NC 3.0
*3:オートエンコーダ(Autoencoder)とは|意味、仕組み、種類、活用事例を解説
*4:CDDD文献中にも引用されています
*5:ちなみのコードのライセンスはApache-2.0です。但しcddd_server.pyもCC BY-NC 4.0なので注意が必要です。
*6:NVIDIA、macOSのサポートを正式に終了した「CUDA Toolkit v11」をリリース。MacではCUDAアプリの開発と実行は出来ない状態に
*7:ネットワークの学習も自分でやろうとしたら不具合が出るのかもしれません。いつも通り無理矢理押し切っているのでおすすめしません。
タンパク質構造のまとめサイト⁉︎ 3D-Beacons Networkで遊んだ話
AlphaFold2、AlphaFold DBの公開から4ヶ月ほどですが、まさに日進月歩という感じで次々と新しいニュースを目にしますね。
複合体予測のAlphaFold-Multimer*1や、Baker研からRosettaFoldと組み合わせた大規模な複合体予測結果*2、さらにAlQuraishi研究室からPyTorch版 OpenFoldが公開されました*3。ますます広く多様な応用が出てきそうですね。
実験によるタンパク質構造解析も「え、そんな構造とけるんだ!複合体すぎるやろ!」みたいな研究がたくさん報告されてびっくりです。*4
発展著しいタンパク質構造解析&予測の分野ですが、専門外の素人からすると「色々ありすぎてよくわかんない。まとめて見られるようにして!」ってなります。
そんな我々に朗報!素敵なまとめサイト(?)が公開されました!
その名も3D-Beacons Network!!

実験データも予測モデルもタンパク質3D構造ならまとめて探せてしまうぜ!しかも安心のEMBL-EBI運営。。。AlphaFold DB公開元なら信頼して遊べるね!
ってなわけで遊んでみましょう!
3D-Beacons Networkって?
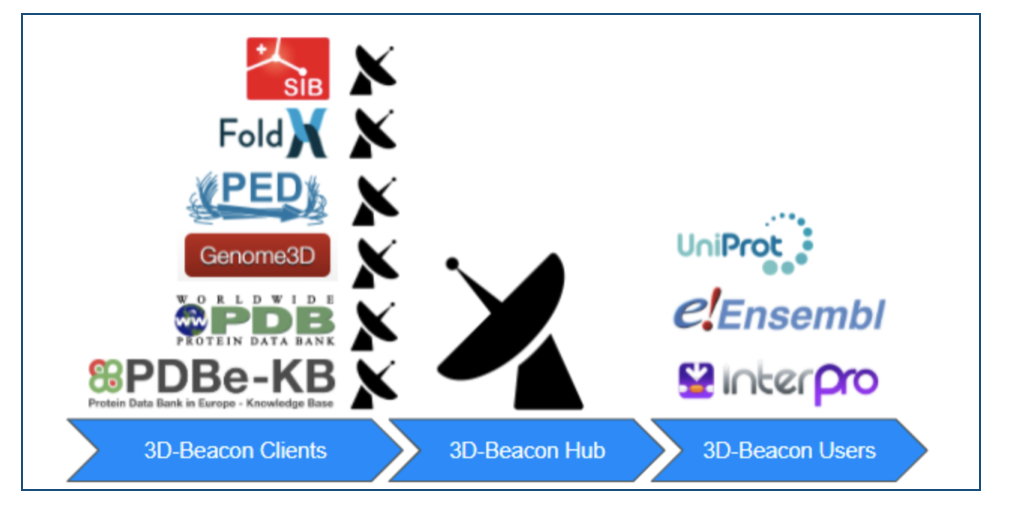
3D-Beacons Networkは複数のリソースから公開されている実験で決定されたタンパク質構造と予測モデルをまとめて一箇所で無料で利用できるようにしたものです。WebページだけでなくAPIも用意されているので、簡単なコードだけでプログラミングによる情報収集・解析も可能です。

データの提供元はこんな感じ。*6
- Protein Data Bank in Europe (PDBe)
- AlphaFoldDB
- SWISS-MODEL
- Protein Ensemble Database (PED)
- Small Angle Scattering Biological Data Bank (SASBDB)
- Genome3D
- PDBe Knowledge Base (PDBe-KB)
これらのデータが統一フォーマットで参照できます。PDBとAlphaFoldしかわからない。。。
Webページをみてみよう
ではさっそく3D-Beacons NetworkのWebページにアクセスしてみましょう。

使い方は簡単!検索したいタンパク質のUniProt IDを検索窓に入れて「Search」するだけです。試しにヒトのEGFR(Epidermal growth factor receptor)「UniProt ID: P00533 (EGFR_HUMAN)」をいれてみました*7。
こんな結果になりました。

実験データに基づく構造が235個、テンプレートに基づくモデルが3個、ディープラニングのモデルが1個の計239構造が登録されているようです。
どんな構造があるでしょうか?
検索結果のページにインタラクティブな3Dビューワーも用意してくれています。さすがEMBL-EBI。

上の図は「PDB ID : 5ug9」が表示されている様子です。「Click to download」をクリックすれば構造データファイルをすぐに取得できます。ここではcifファイルでした。
個人的に便利だと思ったのは3Dビューワーの下にある各構造エントリーとそれぞれがカバーする配列範囲のグラフです。興味のある部分がそのエントリに含まれているか一目瞭然です。

AlphaFold DBがとにかく配列全長をつっこんでいるのが潔くて良いですね!
気になった構造があればエントリー横の目のアイコンをクリックすれば画面上部の3Dビューワーに表示させられます。また、先と同様に下矢印アイコンでダウンロードすることもできます。

またマウスオーバーすると以下のように配列範囲や分解能といった情報も見られます。もちろん元のデータベースへのリンクもあるので、移動して詳細を確認することもできます。

EGFRの例で言えば抗EGFR抗体との相互作用に興味があれば細胞外ドメインを含む構造をしたり、キナーゼ活性部位・低分子阻害剤との相互作用に興味があれば細胞内ドメインを含みそうな構造をチェックしたり、といった遊び方がありそうですね。
APIをつかってみよう
Webページの使い勝手が大体わかりました。Web版はエントリをそれぞれチェックするには便利ですが、「まとめてデータを取得したい!」とか「まとまったデータを別の解析につかいたい!」というときには手間です。
「退屈なことはPythonにやらせよう」ということでAPIをさわってみたいとおもいます。*8
3D Beacons HUB APIのページはこんなでした。

URLを指定してGETすればJSON形式でデータを得られるみたいです。
Pythonならrequestsとかいうのを使えば良いそうです。標準ライブラリではないようですがcondaとかpipで簡単にインストールできます。*9
とりあえずjupyter notebookで試してみます。
# ライブラリのインポート import requests # サーバーURLを指定 Server_url = "https://www.ebi.ac.uk/pdbe/pdbe-kb/3dbeacons/api" # 検索したいUniprot ID(EGFR_HUMAN : P00533) Uniprot_number = "P00533" # APIに従ってURL全体を指定 url = Server_url + "/uniprot/summary/" + Uniprot_number + ".json" # requestsでURLにアクセスしてデータを取得 response = requests.get(url) # HTTPステータスコードの確認 print(response.status_code) # > 200と出力された
HTTPステータスコード 200となりました。無事URLヘのアクセスが成功してデータを取得できたようです。
responseオブジェクトに返ってきているJSONデータはrequestsのjson()メソッドで簡単に確認できるそうです。
json_data = response.json()

情報が多すぎてそのまま表示してもよくわかりませんね。
print(type(json_data)) # <class 'dict'> print(json_data.keys()) # dict_keys(['uniprot_entry', 'structures'])
json()メソッドで変換したデータは辞書型で、keyとしてuniprot_entry、structuresの2つがあるようです。ここで興味があるのは3D-Beacons Networkに含まれている構造情報なので、structuresのvalueを確認すれば良さそうです。
PandasのDataFrameにすれば見やすそうです。
# 辞書型からstructuresのkeyに対応するvalueを取り出す structures_data = json_data['structures'] # PandasのDataFarameに変換 import pandas as pd df = pd.DataFrame(structures_data) df.head()

上図のようなDataFrameができました。「model_identifier」がPDB ID等に相当しそうなので、各行がそれぞれの構造エントリになっているようです。
データ全体を確認してみます。
print(df.shape) # (239, 16)
全239行のDataFrameでした!3D-Beacons NetworkのWebページでEGFRを検索した結果の構造も239エントリーだったのでうまくデータ取得できているようです。
このデータ全体をcsvで出力しておくとプログラミング苦手な私でもExcelなどで気楽に眺められそうです。
df.to_csv("P00533_3D_Beacons.csv", sep=",")
エントリーごとに含まれてる情報はこんな感じ。DataFrameのcolumn名です。
print(df.columns) # Index(['model_identifier', 'model_category', 'provider', 'created', 'sequence_identity', 'uniprot_start', 'uniprot_end', 'resolution', 'coverage', 'model_url', 'model_format', 'experimental_method', 'model_page_url', 'confidence_version', 'confidence_avg_local_score', 'confidence_type'], dtype='object')
model_categoryに「実験によるデータか?予測モデルか?」、model_URLに「データ元のURL」、model_formatに「ファイルフォーマット」といった情報が記載されているようです。
構造データの元ファイルをまとめて取得できるかやってみましょう。
URLから情報を取得するにはライブラリurllibのrequestに含まれるurlopenを使えば良いようです。*10
DataFrameの上から5つを対象にしてみます。
import urllib, time for i in range(5): # IDとフォーマットを使ってダウンロード後のファイル名にする model_ID = df.at[i, "model_identifier"] model_format = df.at[i, "model_format"] if model_format == "MMCIF": extension = ".cif" elif model_format == "PDB": extension = ".pdb" file_name = model_ID + extension # ダウンロードするファイルのURL model_url = df.at[i, "model_url"] # urlretrieveでURLからデータをダウンロードしファイル名をつける model_data = urllib.request.urlopen(model_url).read() with open(file_name, mode="wb") as f: f.write(model_data) # サーバーに負担をかけすぎないようにちょっと待つ time.sleep(20)
無事ファイルが5つダウンロードできました!
上ではDataFrameの各エントリーのmodel_urlにアクセスしてデータを取得しているだけですが、ファイル名をわかりやすくするためにmodel_identifierと、拡張子(extension)の設定にmodel_formatの情報をつかっています。
ついでに連続でアクセスして公共データソースに迷惑をかけないようにちょっと待ち時間(20秒)をはさんでいます。これであっているかはわかりません。すみません。
PyMolでファイル5つを表示してみました。alignするとこんな感じ。

うまく重なっているので目的のタンパク質の構造が取得できていそうですね。
おわりに
以上、実験データと計算による予測モデルの両方を横断したタンパク質立体構造のデータベース3D-Becons Networkで遊んでみたお話でした。
直感的に利用できるWebページが良いですね。また、APIが用意されているのでプログラミングが得意な方であれば、統一フォーマットの情報を色々な用途に利用できるのではないかな?と思います。
私は見様見真似でrequestsを使ってみましたが全く自信がないです。サーバーURLの指定とかアクセスに時間を置くやり方とか間違っていたらごめんなさい。
相変わらず他にもいろいろと間違いが多そうなので、正しい使い方を教えていただけると嬉しいです。
余談①
ところでUniProt IDを使ってPDBとAlphaFold DBのデータをまとめて見るだけなら、UniProtのStructure項でも確認可能です。
ですが、AlphaFold2公開以来のタンパク質構造予測分野の発展のスピードをみていると、これからもどんどんと新しいモデル、データベースが増えていくこと予測されます。
3D-Beacons Networkの利点は統一のフォーマットが用意されており、ソースコードも公開されているため、誰でも3D-Becons Registoryに沿った形にデータを整えて共有できる拡張可能性にあることではないかな?と思います。
また、運営元がEMBL-EBIなので、作っておしまいではなく継続的な運営・発展が期待できるのも魅力かな?とおもいます。
このあたり実際にタンパク質科学に携わられている専門の先生方のご意見を伺いたいところです。
余談②
余談ついでに、先日のサイエンスアゴラ2021でERATO胡桃坂クロマチンアトラスプロジェクトセッションを拝見しました。
クライオ電子顕微鏡を用いたRNAポリメラーゼによる転写メカニズムの解析が非常に印象的でした。
Science文献へのリンク www.science.org
ライフサイエンス統合レビュー 著者による日本語解説
「クロマチンにおいてヒストンに硬く巻きついたDNAをどのようにほどきながら読み取っているか?」という基本的な生命現象がそもそも最近になるまで分かっていなかったということ、また、そのメカニズムがcryoEMでスナップショットをとるように明らかにされていく様子にタンパク質構造解析の威力を感じました。
CryoEMの威力もあってか、どんどん複雑な複合体構造のデータが出てきて、またAlphaFoldやRosettaFoldによる複合体予測も公開されてきています。複合体に割り当てるIDはどうするんだろう?など素人的には気になるところです。
ますます発展する構造解析・予測の分野の共通のインフラとして3D-Beacons Networkがこの先どのように使われていくかも楽しみですね。
*1:https://www.biorxiv.org/content/10.1101/2021.10.04.463034v1
*2:https://www.science.org/doi/10.1126/science.abm4805
*3: OpenFoldには公開版AlphaFold2にはなかったトレーニング部分も含まれているそうです GitHub、colab
*4:10年前はGPCRが一つ公開されると異分野の私でもニュースを見るくらい大変な大騒ぎだった気がするのに。。。科学の発展はすごいです。
*5:図中のいらすとは「いらすとや」さんから、ロゴは各サイトのロゴを利用させていただいています
*6:EMBL Newsより 3D-Beacons Network: protein structure data, all in one place
*7:EGFR_HUMANのUniProtページはこちら UniProtKB - P00533
*8:そんな名前の書籍があった気がしますが未読です。すみません。
*9:note.nkmk.me 「Python, Requestsの使い方」
深層学習っぽい用語のメモ 〜 Auxiliary loss、Transformer-XL、axial-attention〜
AlphaFold2はディープラーニングの専門家の視点でみても面白いそうですが、ど素人にはさっぱりです。 というわけで、前回の「Self distillation」に引き続き、深層学習っぽい用語を調べています。
今回取り上げる用語はAuxiliary loss、Transformer-XL、axial-attentionです。
概要
むりやり1枚にまとめるとこんな感じの話です。

初めに青枠の①auxiliary lossをみます。損失計算における工夫で、勾配消失といった課題と関わりそうです。
次に緑枠、Transformerの固定長という制限を取り除こうとする取り組みをみます。auxiliary lossを応用した②vanilla modelや、その改良版③Transformer-XLです。
最後に赤枠、画像認識へのSelf-attentionの応用(④)をみます。改良版として、2Dデータの処理を軸方向の1Dの組み合わせに落とし込んだ⑤axial-attentionがでてきます。Transformer-XLで出てくる相対的位置符号化の考え方はこっちにも顔を出します。*1
では順番に見ていきましょう!
1. Auxiliary loss
1-1. こまめな中間報告が成功の鍵? ~補助損失~
「Auxiliary loss(補助損失)」は2014年の画像認識のコンペティション ILSVRC(ImageNet Large-Scale Visual Recognition Competition)で優勝したモデル GoogLeNet で利用され、有名となった手法のようです。
- 文献①: C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich.
Going deeper with convolutions. In Proc. of CVPR, 2015. arXiv.:1409.4842v1
GoogLeNetは下図のようなアーキテクチャーのネットワークです。めちゃくちゃディープで複雑なので途中で分割してます。
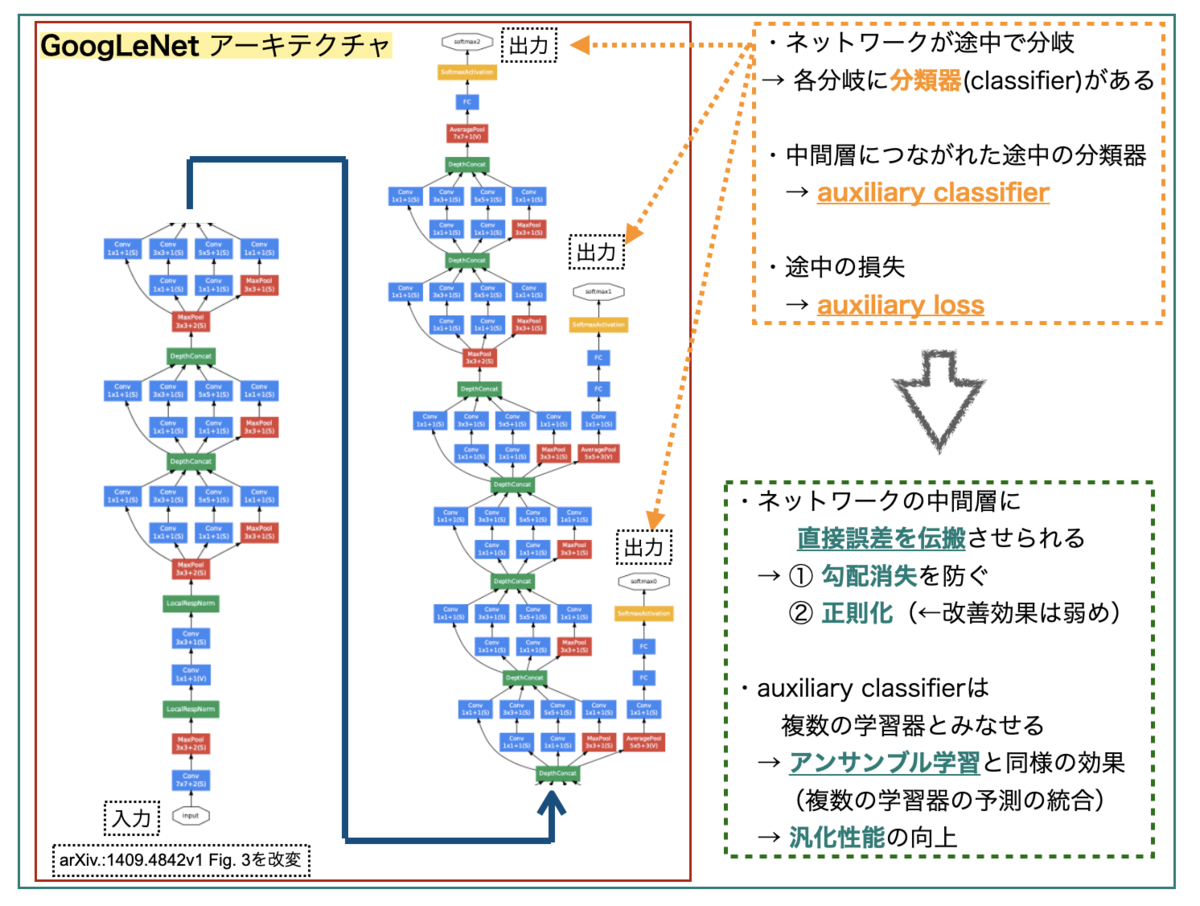
GoogLeNetの特徴の一つは、ネットワークが途中で分岐してサブネットワークを構成していることです。*2ネットワークの最後だけでなく、各サブネットワークにもそれぞれクラス分類器(classifier)があり、auxiliary classifierと呼ばれています。 これらから求められる損失がauxiliary lossとなります。
巨大なネットワークでは「全ての層に勾配を効率的に伝搬するのが難しい」という課題があります。GoogLeNetでは、中間層にauxiliary classifierが繋がっていることで、中間層に直接誤差を伝搬することができ、① 勾配消失を防ぐ、② 正則化を実現できる*3、といった利点があるそうです。*4
また、複数のクラス分類器があるので、アンサンブル学習(複数の学習器の予測結果を統合)と同様の効果が得られ、汎化性能を高めることが期待できるそうです。*5
巨大なモデルの途中途中で、評価とフィードバックをうけるとうまくいく、って感じでしょうか?深層学習でもこまめな進捗の報連相、大事なんですねー。
1-2. AlphaFoldででてくるAuxiliary loss
AlphaFold2では学習に使う損失関数として、タンパク質立体構造予測のための特別な「FAPE (Frame Aligned Point Error)」というオリジナルな損失を作成しています。 さらに学習では、FAPEに加えて多数の「補助的な(Auxiliary)損失」を合わせて使っています。

上図のL_auxはStructure Moduleで計算されるauxiliary lossです。Structure Moduleは8層(8 blocks)の積み重ねとなっています。 各層ごとに、その時点での中間的な3次元構造について損失を計算し、最後にすべての層での損失の平均を取ってL_auxとしています。
L_auxはFAPE損失とねじれ角損失(torsion angle loss)を足し合わせたものです。この中間的な損失では計算負荷を減らすため、 FAPEは主鎖Cαのみ、側鎖に関する損失はねじれ角のみにする、という工夫が行われているようです。
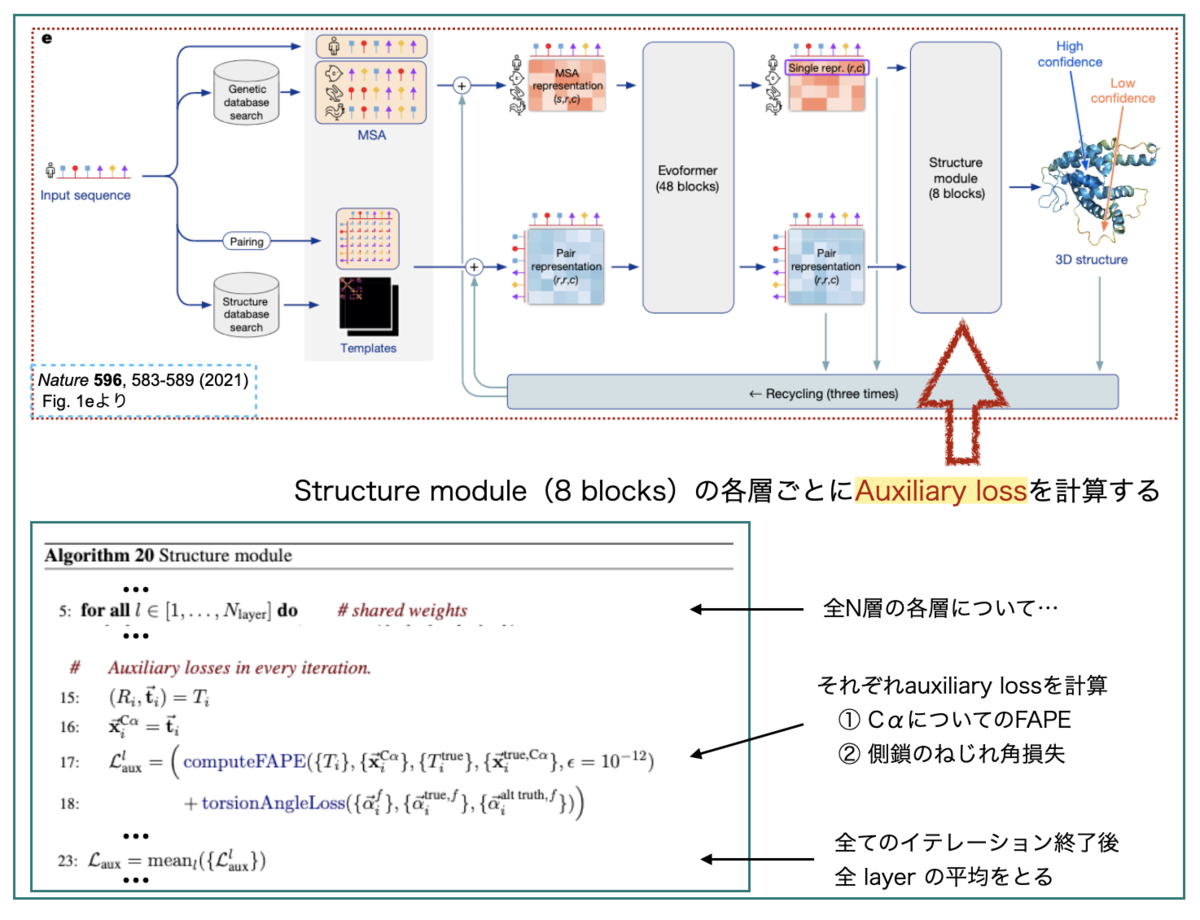
GoogLeNetの時と同様に、途中結果の損失評価を挟むことで、巨大なモデル全体の学習を成り立たせたり、汎化性能を高めたりできてる、ということでしょうか??
1-3. Auxiliary lossの効果はあったの? ~ablation study~
Auxiliary lossでやりたいこととAlphaFold2での使われ方はだいたい分かりましたが、実際のところどれくらい効果があったのでしょうか?
AlphaFold2の文献では評価実験も報告されています。「複雑な構成要素の一部を取り除いた手法」を「オリジナルの手法」と比較する評価実験を ablation study というそうです。*6
本文 Fig. 4a(下図)に結果、Supplementary Information 1.13に詳細な説明が載っています。

異なるベースラインのシード3つで実験した平均で比較しているので、各要素それぞれ3本ずつ結果があるようです。
「No auxiliary distogram head」や「No auxiliary maked MSA head」がauxiliary lossに関連する項目となりそうでしょうか? 概してベースライン(縦線)より小さくなっているので、「元々のモデルでは精度に貢献していた」ということになりそうです。
Supplementary Informationのコメントを引用しておきます。
Finally, careful handling of the structure and intermediate losses (using both end-to-end structure gradients and providing intermediate loss gradients in recycling) is important to achieving full accuracy. This is likely due to pushing the network towards having a concrete representation of the structure as appears to be present in the trajectories of intermediate structure predictions.
(Supplementary Information p51 強調は追加)
良かった良かった(?)
2. Transformer-XL
次の用語はTransformer-XLです。XLはextra longの意です。
- 文献②:Z. Dai, Z. Yang, Y. Yang, J. Carbonell, Q. Le, and R. Salakhutdinov.
Transformer-XL: Attentive language models beyond a fixed-length context. arXiv:1901.02860v3
AlphaFold2はTransformerをベースにしたモデルですが、Trasformerには「固定長しか扱えない」という制限があるそうです。これをうまく克服したのがTransformer-XLで「セグメントレベルの再起」と「相対位置符号化」がポイントだそうです。 *7
Transformer-XLの論文の中身を見る前に、論文中で比較されている以前のモデルを見ておきましょう。
2-1. Transformer x Auxiliary lossで文字レベル言語モデルの高性能化
- 文献③:R. Al-Rfou, D. Choe, N. Constant, M. Guo, and L. Jones.
Character-level language modeling with deeper self-attention.arXiv:1808.04444v2
文献③は、LSTMやRNNが高性能を発揮していた「Character-Level Language Modeling」でも、Transformerを使うことでより高い性能が出せることを示した文献です。
Character-Level Language Modelingは、「単語(word)レベル」ではなく「文字(character)レベル」の言語モデルのことで、ここで扱われているのは「入力の文字列から次の文字を予測」するタスクです。*8
文献③ではディープ(64層)なTransformerモデルを使っていますが、成功の鍵として「中間層および配列の中間位置にauxiliary lossを加えたこと」を上げています。これにより「収束のスピード」を上げることができ、より深いネットワークの学習ができたそうです。
モデルの構造をステップ・バイ・ステップでみるとこんな感じです。中間位置、中間層、複数ターゲットの予測とタスクを増やして、それぞれで補助損失(auxiliary loss)を計算するという構造になっています。

また、「文字の位置情報の取り扱い(Positional embedings)」も工夫しています。
深いモデルでは、入力の段階にだけ位置情報を付け加えても、層間の伝搬中に情報が失われる可能性があります。そこで、各transformer層に入る前の入力配列に、層毎に学習する「位置の埋め込み(positional embedding)」を加えています。
以上のようなモデルにより高い精度を発揮することができたとのことです。Auxiliary lossがここでも活躍していますね。
くどい感じに図を引用しましたが、この図の見方がわかるとTransformer-XLの図が少し分かりやすくなります。
2-2. Transformer-XLと可変長文字列の取扱い
Transformer-XLの利点は「可変長を扱える」ことで、ポイントは「① セグメントレベルの再起」と「② 相対的位置符号化」とのことでした。順番にみていきます。
2-2-1. セグメントレベルの再起(Segment-level Recurrence)
まず①について、比較対象として「(再起のない)セグメント化モデル」があげられています。これが先の文献③です。
可変長を扱うには、「どうやって長文を固定長の表現に符号化(encode)するか?」が課題となります。単純に「制限のないTransformerを用意して全文を投げ込む」のでは膨大な計算リソース必要で現実的ではありません。
文献③のアイデアは「コーパス全体を、扱いやすいサイズの短いセグメントに分割し、各セグメント内部でだけ訓練を行う」というものです(文献②ではvanilla modelと呼んでいます)。このモデルでは、セグメント間での情報のやりとりはありません。
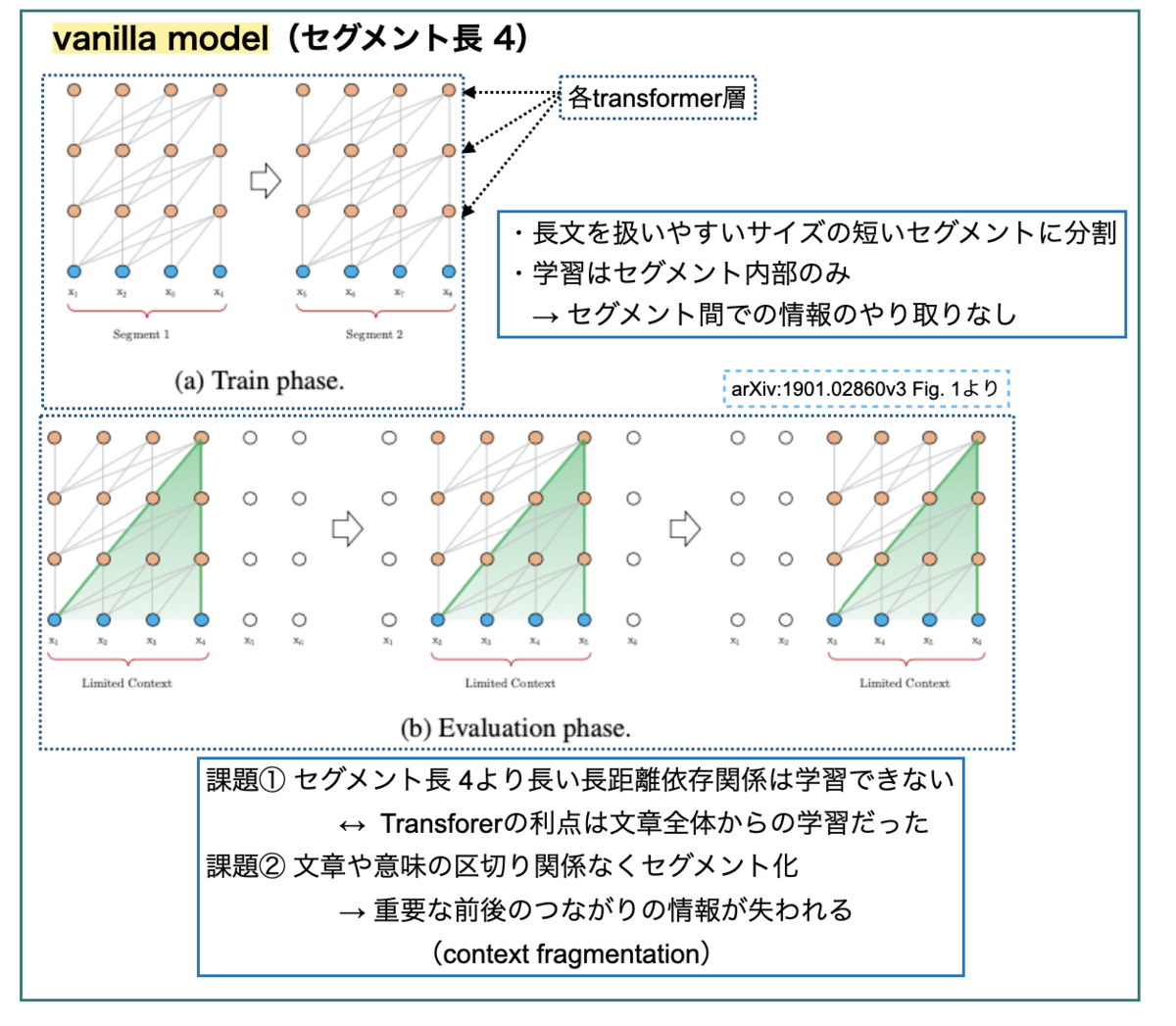
vanilla modelの限界として2点指摘されています。
- 決まった長さ以上の長距離依存関係が学習できない(上限がセグメントの長さ)
- 文章や意味の区切りを考慮せずにセグメントに切り分けることで重要な情報が失われる(context fragmentation)
以上の課題を解決するため、Transformer-XLは「再起メカニズム(recurrence mechanism)」を導入しています。文献② Fig.2 を見るとイメージしやすいです。

前のセグメントの「隠れ状態配列(hidden state sequence) 」を計算し、固定化・キャッシュ化します(Fixed(No Grad))。これを次の新しいセグメントを処理する際に「拡張文脈(extended context)」として再利用します。
このように「セグメントレベルで再起処理」を行うことで、セグメント境界を超えた情報の伝達が可能になり、長距離依存関係の学習ができたり、context fragmentationが解消できる、ということのようです。
2-2-2. 相対的位置符号化(Relative Positional Encoding)
文献②(vanilla model)で位置情報の取扱いについてふれましたが、Transformer-XLでは「隠れ状態」を再利用するようにしたことで新たな問題が生じました。
標準的なTransformerでは、位置情報は「絶対的(absolute)な位置」として表せば十分で、これはセグメント内部でしか情報をやりとりしない vanilla modelでも一緒です。
一方、セグメント間のやり取りのあるTransformer-XLで「セグメント内の絶対的位置」([0, 1, 2, 3])を使ってしまうと、「隠れ状態(前のセグメント)の内部における位置」との区別ができなくなってしまいます([0, 1, 2, 3, 0, 1, 2, 3])。
そこで「隠れ状態の「相対的(relative)な位置情報」だけをエンコードするようにしよう!」というのが「相対的位置符号化」です。
TransformerのAttention操作について考えると、計算するだけならqueryとkeyの「それぞれのベクトル内部での絶対的位置(i、j)」がわからなくても、「相対的な距離(i-j)の情報」さえあれば十分計算できるよね、ってことみたいです。
なお、相対距離がわかっていれば、後から再起的に絶対的な位置情報を取得することができるそうなので安心(?)ですね。
「Relative positional encodingを実際にどのように行うか?」は文献③中、また以下の記事で丁寧に解説してくださっています。*9
2-3. AlphaFold2と相対的位置符号化
AlphaFold2でTransformer-XLが引用されている箇所は「Supplementary Information 1.5 Input embeddings」です。
ペア表現(pair representation)について、鎖(chain)における残基の位置の情報をネットワークに与える際に「相対的な位置の特徴(relative positional features)」をエンコードしています。
具体的には、Algorithm 4のように、最大値 32で区切ってone-hotベクトル表現としています。相対的距離(差)で表すやり方がTransformer-XLに類似しているとのようです。

「実験的には、相対的なエンコードにより、学習に使った配列よりも長い配列に対しても質を落とさずにネットワークを評価することができる」とのことですが、このあたりTransformer-XLのEvaluation Phaseでセグメントを超えた情報伝達が行われている、という点と関連があるのでしょうか??
2-4. 相対的位置符号化とAlphaFold2の複合体予測
可変長を扱えるTransformer-XLについて、AlphaFold2では「相対的位置符号化」の技術が転用されているようでした。この技術について、AlphaFold2をGoogleColab上で使えるようにし、さらに機能の拡張をおこなったColabFold論文で面白い指摘がされていました。
- 文献④: M. Mirdita, S. Ovchinnikov and M. Steinegger
ColabFold - Making protein folding accessible to all bioRxiv 2021.08.15.456425
先に見た通り、AlphaFold2では「 |i-j|≧ 32でキャップした相対的位置符号化」を使っています。従って、32以上離れた残基のペアはどれでも、同じ相対的位置符号化がなされることになります。 つまりAlphaFold2は、2つのタンパク質間の残基のインデックスを32よりも大きく離せば、それぞれ別々のポリペプチド鎖として扱う、といことになります。
これを踏まえて、AlphaFold2でタンパク質-タンパク質複合体予測を行いたい場合には、「それぞれの鎖の間に32残基分より大きく間隔をあけてつないで入力すれば良い」らしく、ColabFoldにおける複合体予測機能の実装に利用しているそうです。
「なぜAlphaFold2で複合体予測がうまくいくのか?」という問いの答えにはなっていませんが、モデルの仕組みを利用して新しい機能の実装につなげているのは面白いですね。
3. axial-attention
最後に取り上げる用語はaxial-attention(もしくはcriss-cross atention)です。
元々は、「画像認識の分野Attentionを使いたい」というところから来ているようです*10。ざっくり「2次元データの各軸方向にattentionを使うぜ!」って感じみたいです。
AlphaFold2では、本文(ref. 53)とSupplementary Information(ref. 118)にそれぞれ関連する文献が引用されています。
- 文献⑤: H. Wang, Y. Zhu, B.Green, H. Adam, A. Yuille and L.-C. Chen
Axial-DeepLab: Stand-Alone Axial-Attention for Panoptic Segmentation arXiv:2003.07853v2 - 文献⑥: Z. Huang, X. Wang, Y. Wei, L. Huang, H. Shi, W. Liu and T. Huang
CCNet: Criss-Cross Attention for Semantic Segmentation arXiv:1811.11721v2
3-1. 画像認識へのAttentionの利用
以下の記事がとても分かりやすかったです。
単純に画像認識のConvolutionをSelf-Attentionに置き換えた初期の例として以下の文献があるそうです。
- 文献⑦: P. Ramachandran, N. Parmar, A. Vaswani, I. Bello, A. Levskaya and J. Shlens
Stand-Alone Self-Attention in Vision Models arXiv:1906.05909v1
畳み込みの空間領域に局所的なattention層を当てはめています。こういう感じ。

位置情報は、相対的距離(relative distance)を利用して取り込んでおり、relative attentionと呼んでいます。

Transformer-XLを見た後だとやりたいことが分かりやすいですね。
3-2. 軸毎に切ったattentionで画像の広範囲を取り込む ~axial-attention~
画像認識に対してAttentionを使う雰囲気は分かりましたが、先の例は局所的な領域(ex. 3x3)への適用でした。 元々のTransformerに「自然言語処理で文章全体からの情報を利用したい」というモチベーションがあったことを考えると、画像認識でも同様に「範囲をより広く全体に広げたい」という気持ちになりそうです。
これは画像認識のパノプティックセグメンテーションというタスクと関係してくるようです。*11

上のようなタスクを行うには、モデルが認識できる画像の範囲(受容野, receptive field)を広くとって、局所的ではなく大域的な空間関係を把握することが大事となってくるようです。
画像認識のAttentionでも範囲を広げたくなりましたが、2次元で領域を単純に広げるのは計算コストがかかりすぎて難しいそうです。そこで「2次元ではなく1次元で軸方向にattentionを使うことでこの問題を回避しよう」というのが文献⑤です。*12

上図のように、「高さ軸方向(hight-axis)」と「幅軸方向(width-axis)」の1D attentionを順次適用しています。これにより、2D attentionよりも計算効率を改善し、大域的な情報(広い受容野)を使用することができるようになった結果、パノプティックセグメンテーションのタスクで良い結果を示したそうです。
また、axial-attentionでは文献⑦よりも「相対的位置符号化(relative positional encoding)」を拡張していて、keyだけでなく、queryとvalueにもpositional biasを加えています。

以上がaxial-attentionの概要でした。
3-3. AlphaFold2でのaxial-attention
AlphaFold2でaxial-attentionが出てくるのは、EvoformerのMSA部分です(Supplementary Information 1.6 Evoformer blocks)。 入力の多重配列アラインメント(MSA)に対して、行・列それぞれについて「MSA row-wise gated self-attention」(SI 1.6.1)と「MSA column-wise gated self-attention」(SI 1.6.2)を利用しています。

また、Ablation Studyの一つ「No triangles, biasing, or gating (use axial attention)」(SI 1.13.1)では、row-wise MSA attentionのペアバイアス(pair bias)を「投影された相対的距離(relpos_ij)」と置き換えた後、すべてのattention操作からゲートを取り除いたモデルを作成しています。
このような修正を加えてシンプルにしたものは、文献⑥に出てくるような「標準的なaxial attention(あるいはcriss-cross attention)」とほぼ似たような構造となっているそうです。
4. まとめ
以上、今回は前回に引き続きAlphaFold2に出てくる深層学習っぽい用語、Auxiliary loss、Transformer-XL、axial-attentionについてでした。
色々なテクニックを取り入れられて作られているとは、方々で目にしていましたが、調べれば調べるほど色々なトピックがでてきてびっくりしています。「この単語1語の背景に先行研究がたくさんあったのか!」の連続です。
また、用語を検索したら日本語で解説してくださっている記事が出てきて、「インターネットの賢い人たち本当にありがとう」と感謝しきりでした。参考にさせていただいた記事は全て本文と脚注に記載したつもりですが抜けていたら教えていただけると嬉しいです。
深層学習の用語というわけではありませんが、「Ablation Study」がスタンダードな評価実験の方法、というのもつでいに分かって、AlphaFold2文献の中で意味のわかるFigureが少し増えました。Figureに書かれている効果の大きさがどれくらいなのかはピンときませんでしたが。。。
以上、今回も適当に切り貼りした記事になってしまいました。間違いがとても多そうなのでご指摘いただければ幸いです。ではでは。

*1:研究の時系列とは関係なく、今回の記事のお話の流れです。すみません
*2:他にもInceptionというモジュールを使っていることなど、魅力的な特徴はいっぱいあるそうですがここでは割愛します。
*4:参考: 内田 祐介 山下 隆義「物体認識のための畳み込みニューラルネットワークの研究動向」電子情報通信学会論文誌D Vol.J102-D No.3 pp.203-225
*5:ディープラーニングのE資格の例題としてでてきました。 参考: Qiita DNN_後編2 (確認テストの考察)
*6:参考:「研究における評価実験で重要な7つのこと」
*7:参考①:Google AI Blog 解説記事「Transformer-XL:Unleashing the Potential of Attention Models」
参考②:WebBigDataさんによる参考①記事の翻訳 「Transformer-XL:Attentionモデルの可能性を解き放つ」
参考③:楽しみながら理解する自然言語処理入門「Transformer-XLを理解する」
*8:文字を扱う「Character-Level Language Modeling」は以下の点で難しいそうです。
1. 単語の語彙を「1から」学習しないといけない
2. 自然言語のテキストは何百・何千ものステップを介した長距離依存を示す
3. 文字列は単語列よりも長くなるので計算のステップ数がかなり多くなる。
*9:私にはさっぱりわからなかったです。
*10:参考:Qiita「画像認識でもConvolutionの代わりにAttentionが使われ始めたので、論文まとめ」
*11:参考:SkillUp AI 深層学習を用いたセグメンテーションの紹介 セグメンテーションシリーズ①
参考文献:A, Kirillov, K.He, R. Girshick, C. Rother and P. Dollár Panoptic Segmentation arXiv:1801.00868ve
*12:参考①:Google AI Blog 解説記事「Axial-DeepLab: Long-Range Modeling in All Layers for Panoptic Segmentation」
参考②:WebBigDataさんによる参考①記事の翻訳 「Axial-DeepLab:パノプティックセグメンテーション用にattentionを改良(1/2)」
「Axial-DeepLab:パノプティックセグメンテーション用にattentionを改良(2/2)」